You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001261_03414
You are here: Home > Sequence: MGYG000001261_03414
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Caulobacter sp903900155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; Caulobacter sp903900155 | |||||||||||
| CAZyme ID | MGYG000001261_03414 | |||||||||||
| CAZy Family | PL10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3229; End: 4824 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL10 | 116 | 384 | 2.8e-128 | 0.9745454545454545 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02474 | pec_lyase | 4.68e-23 | 116 | 297 | 1 | 182 | pectate lyase, PelA/Pel-15E family. Members of this family are isozymes of pectate lyase (EC 4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. [Energy metabolism, Biosynthesis and degradation of polysaccharides] |
| pfam09492 | Pec_lyase | 1.42e-22 | 116 | 297 | 1 | 180 | Pectic acid lyase. Members of this family are isozymes of pectate lyase (EC:4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. |
| pfam09492 | Pec_lyase | 4.64e-08 | 49 | 209 | 3 | 158 | Pectic acid lyase. Members of this family are isozymes of pectate lyase (EC:4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. |
| cd02889 | SQCY | 4.55e-05 | 85 | 355 | 99 | 341 | Squalene cyclase (SQCY) domain; found in class II terpene cyclases that have an alpha 6 - alpha 6 barrel fold. Squalene cyclase (SQCY) and 2,3-oxidosqualene cyclase (OSQCY) are integral membrane proteins that catalyze a cationic cyclization cascade converting linear triterpenes to fused ring compounds. Bacterial SQCY catalyzes the convertion of squalene to hopene or diplopterol. Eukaryotic OSQCY transforms the 2,3-epoxide of squalene to compounds such as, lanosterol (a metabolic precursor of cholesterol and steroid hormones) in mammals and fungi or, cycloartenol in plants. Deletion of a single glycine residue of Alicyclobacillus acidocaldarius SQCY alters its substrate specificity into that of eukaryotic OSQCY. Both enzymes have a second minor domain, which forms an alpha-alpha barrel that is inserted into the major domain. This group also contains SQCY-like archael sequences and some bacterial SQCY's which lack this minor domain. |
| cd02892 | SQCY_1 | 5.14e-05 | 90 | 209 | 387 | 499 | Squalene cyclase (SQCY) domain subgroup 1; found in class II terpene cyclases that have an alpha 6 - alpha 6 barrel fold. Squalene cyclase (SQCY) and 2,3-oxidosqualene cyclase (OSQCY) are integral membrane proteins that catalyze a cationic cyclization cascade converting linear triterpenes to fused ring compounds. This group contains bacterial SQCY which catalyzes the convertion of squalene to hopene or diplopterol and eukaryotic OSQCY which transforms the 2,3-epoxide of squalene to compounds such as, lanosterol in mammals and fungi or, cycloartenol in plants. Deletion of a single glycine residue of Alicyclobacillus acidocaldarius SQCY alters its substrate specificity into that of eukaryotic OSQCY. Both enzymes have a second minor domain, which forms an alpha-alpha barrel that is inserted into the major domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTC92004.1 | 1.18e-302 | 1 | 526 | 1 | 529 |
| ALL14486.1 | 1.55e-288 | 1 | 526 | 1 | 527 |
| QTN20460.1 | 3.03e-277 | 1 | 527 | 1 | 528 |
| QYF87102.1 | 1.04e-259 | 3 | 531 | 6 | 535 |
| ADL02245.1 | 1.33e-254 | 1 | 527 | 1 | 527 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R76_A | 1.03e-14 | 103 | 310 | 81 | 303 | ChainA, pectate lyase [Niveispirillum irakense] |
| 1GXM_A | 8.58e-12 | 98 | 318 | 26 | 234 | Family10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXM_B Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXN_A Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus] |
| 1GXO_A | 8.73e-11 | 98 | 318 | 26 | 234 | MutantD189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid [Cellvibrio japonicus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as TAT
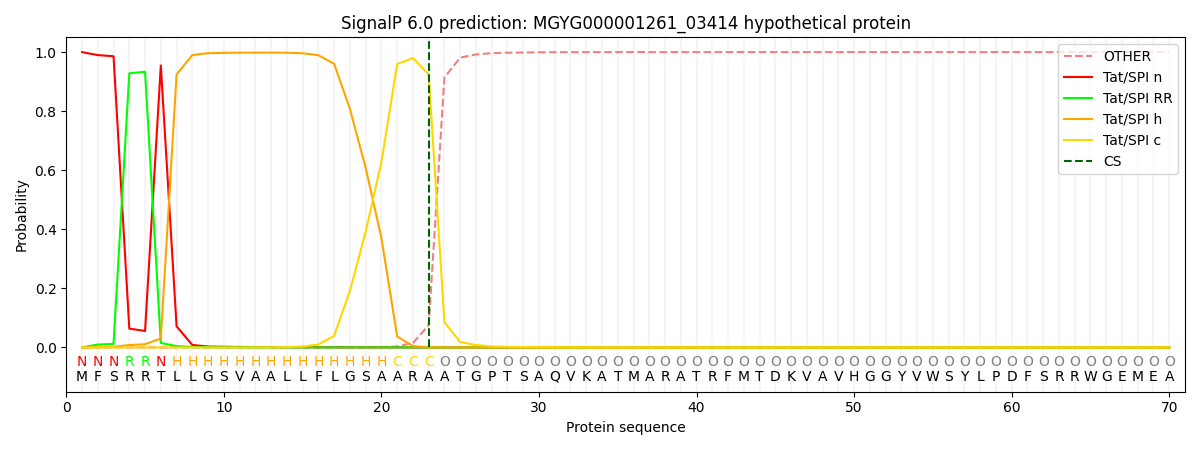
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000008 | 0.000000 | 0.999966 | 0.000007 | 0.000000 |
