You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001254_01931
You are here: Home > Sequence: MGYG000001254_01931
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
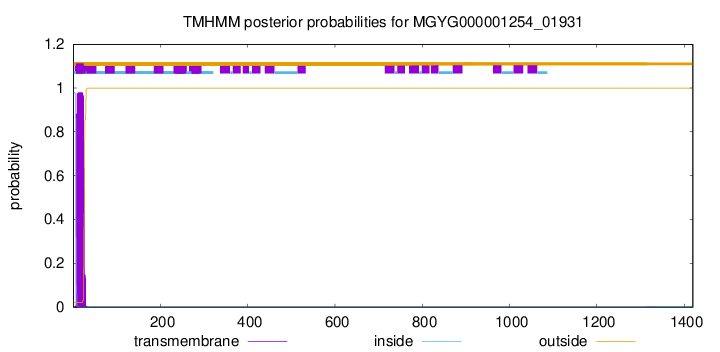
TMHMM annotations
Basic Information help
| Species | TF01-11 sp900759665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp900759665 | |||||||||||
| CAZyme ID | MGYG000001254_01931 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28808; End: 33070 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 351 | 599 | 1.5e-82 | 0.9831223628691983 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.07e-65 | 348 | 606 | 1 | 270 | Cellulase (glycosyl hydrolase family 5). |
| PHA03247 | PHA03247 | 1.21e-11 | 654 | 795 | 2610 | 2767 | large tegument protein UL36; Provisional |
| pfam05109 | Herpes_BLLF1 | 3.70e-11 | 571 | 878 | 336 | 655 | Herpes virus major outer envelope glycoprotein (BLLF1). This family consists of the BLLF1 viral late glycoprotein, also termed gp350/220. It is the most abundantly expressed glycoprotein in the viral envelope of the Herpesviruses and is the major antigen responsible for stimulating the production of neutralising antibodies in vivo. |
| NF033761 | gliding_GltJ | 5.06e-11 | 662 | 795 | 389 | 522 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| PHA03247 | PHA03247 | 1.30e-10 | 653 | 795 | 2736 | 2882 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGH40913.1 | 8.76e-119 | 266 | 642 | 22 | 391 |
| AAC06197.1 | 1.08e-118 | 266 | 642 | 3 | 372 |
| AAC06196.1 | 4.92e-118 | 304 | 659 | 6 | 357 |
| CBK74991.1 | 1.87e-116 | 321 | 660 | 40 | 367 |
| AGH41463.1 | 4.14e-113 | 261 | 659 | 167 | 564 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 2.87e-78 | 325 | 637 | 4 | 296 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZW_A | 1.00e-70 | 326 | 637 | 4 | 298 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 1.03e-70 | 324 | 637 | 2 | 299 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 3PZT_A | 3.44e-67 | 326 | 637 | 29 | 320 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 1LF1_A | 4.30e-64 | 328 | 637 | 6 | 296 | CrystalStructure of Cel5 from Alkalophilic Bacillus sp. [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15704 | 1.09e-68 | 347 | 655 | 54 | 341 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P22541 | 5.50e-66 | 331 | 652 | 113 | 418 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P07983 | 2.48e-65 | 326 | 646 | 34 | 334 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| Q59394 | 1.29e-64 | 322 | 652 | 29 | 337 | Endoglucanase N OS=Pectobacterium atrosepticum OX=29471 GN=celN PE=3 SV=1 |
| P10475 | 2.09e-64 | 326 | 637 | 34 | 325 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
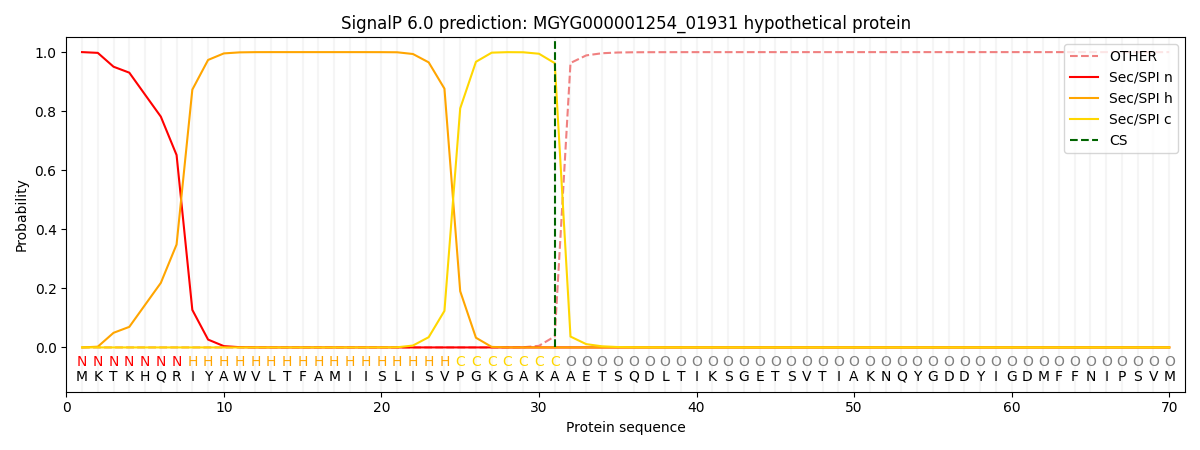
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000451 | 0.998590 | 0.000357 | 0.000194 | 0.000191 | 0.000173 |