You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001183_01846
You are here: Home > Sequence: MGYG000001183_01846
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1441 sp900551755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441; UMGS1441 sp900551755 | |||||||||||
| CAZyme ID | MGYG000001183_01846 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19537; End: 21711 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 58 | 412 | 6.7e-92 | 0.9836956521739131 |
| CBM77 | 616 | 714 | 4.3e-20 | 0.941747572815534 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18283 | CBM77 | 2.14e-20 | 618 | 721 | 6 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| cd14256 | Dockerin_I | 2.75e-08 | 555 | 609 | 1 | 54 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 3.50e-06 | 556 | 609 | 1 | 52 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| COG5434 | Pgu1 | 2.93e-05 | 63 | 397 | 85 | 416 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam13229 | Beta_helix | 4.05e-05 | 241 | 375 | 53 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASW75534.1 | 5.01e-60 | 67 | 520 | 30 | 489 |
| QCX40893.1 | 2.73e-57 | 62 | 520 | 23 | 479 |
| AET61906.1 | 5.43e-57 | 62 | 519 | 38 | 511 |
| AZJ24015.1 | 6.01e-57 | 62 | 379 | 47 | 369 |
| CUU48905.1 | 4.31e-56 | 64 | 521 | 40 | 459 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5FU5_A | 3.27e-11 | 616 | 723 | 9 | 113 | Thecomplexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition [Ruminococcus flavefaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94576 | 6.10e-42 | 61 | 480 | 36 | 454 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
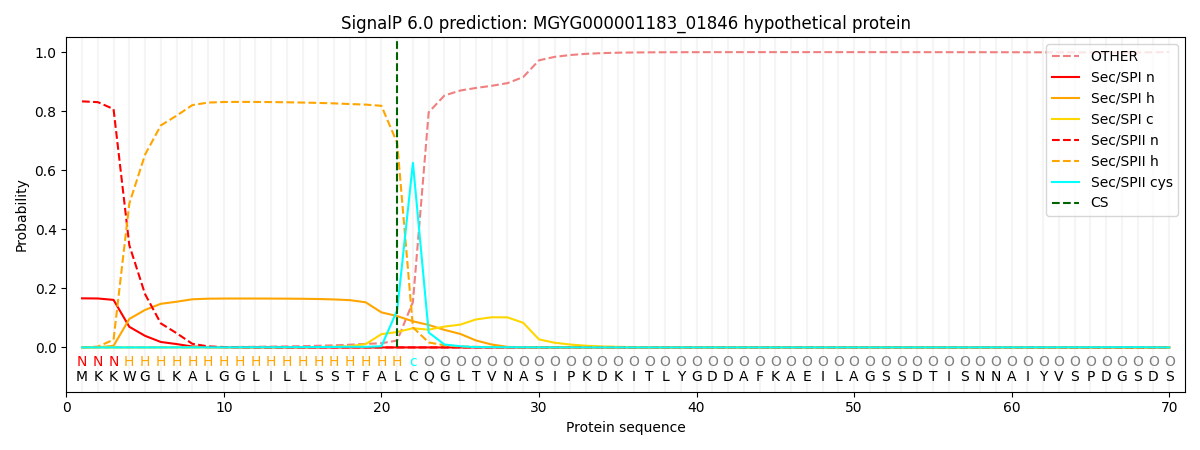
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001021 | 0.161787 | 0.836706 | 0.000175 | 0.000154 | 0.000128 |
