You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001172_00303
Basic Information
help
| Species |
Fusobacterium sp000235465
|
| Lineage |
Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium; Fusobacterium sp000235465
|
| CAZyme ID |
MGYG000001172_00303
|
| CAZy Family |
GT21 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 397 |
|
46796.63 |
9.1648 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001172 |
2065216 |
MAG |
Austria |
Europe |
|
| Gene Location |
Start: 52784;
End: 53977
Strand: +
|
No EC number prediction in MGYG000001172_00303.
| Family |
Start |
End |
Evalue |
family coverage |
| GT21 |
44 |
270 |
3.4e-20 |
0.9699570815450643 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13506
|
Glyco_transf_21 |
1.12e-38 |
99 |
274 |
2 |
174 |
Glycosyl transferase family 21. This is a family of ceramide beta-glucosyltransferases - EC:2.4.1.80. |
| COG1215
|
BcsA |
1.10e-04 |
26 |
384 |
39 |
411 |
Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000051
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
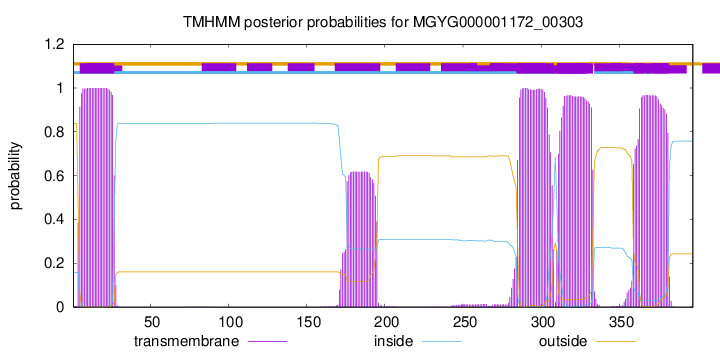
| start |
end |
| 5 |
27 |
| 285 |
307 |
| 311 |
333 |
| 359 |
381 |


