You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001027_00441
You are here: Home > Sequence: MGYG000001027_00441
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900752065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900752065 | |||||||||||
| CAZyme ID | MGYG000001027_00441 | |||||||||||
| CAZy Family | GH101 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5516; End: 12166 Strand: + | |||||||||||
Full Sequence Download help
| MKEKGNKQRL RSFMAALLAL VLAFSPVLSS IPAIQAEAAD AKIWYHVETG SGNGNAHTYS | 60 |
| DASTSPAAVL LHRTATMPAD GTFSVTYEKQ GDNTASARLG FFYTYVDDSN FLYVGFDTQS | 120 |
| HWFYEYKVNG GGSYSTISGL PDQTDGSRTD FSISVSREAL TVKVNDTIKS VNVQDFVSLM | 180 |
| EKAGGNGRFG FRGGNPTSFH FTDAKVNNEA VSNEWGFLAD CAGQTFQEEE PVPVYTVTGT | 240 |
| VKNTANQPIA GANVRIGYSS TKTDAAGSFS IDVEEGTYDV TVSAAGYAAQ TVAGVEINSN | 300 |
| KSLEAMSLVE KAAVVYKNYI QKDSIKAAVS EKFPQVMQYK LNVDGNEKVF AGQEDELSVI | 360 |
| KINGVAITPV MGDVQINADN AVYPMTLKKD SIDMEMTVKI SVKENDLTWE VTDITKKEGC | 420 |
| AKIHTIEVPD LNLVTISDEQ KDAQFMGANV SGNVQVSGDE KITFDEGFSA NSSAGYAYGF | 480 |
| LSADGISAGV WSNSEMAGDK RLVRNNSANS MSLTSALWYY EYGDLAASAT LDGTPVSELP | 540 |
| STKVCLAGDE NRDGTVDWQD GAIAYRDIMN NPYGSENTKD LVNYRISMNF SSQATNPYLK | 600 |
| TADNIKKVYL ATDGLPQAVM MKGYGSEGHD SANSEYGKIA DRLGGLEELK KLNSIAHQYH | 660 |
| TQMGIHINAQ EVYPEAQSFT NELINGPSAL GWGWLDQSYT INRPYDLASG LRYKRLFQLY | 720 |
| DQLNGTSLYA NKWPGVVGTG TDETVADAET IAETVAEKKL TTDENLDFMY LDVWYGDSWE | 780 |
| TRKIAQQINS LGWRFSTEFG YEGEYDSTWQ HWATEGHYGG SSMKGLNSDV IRFIRNHQKD | 840 |
| SFVLNYPYFN NSYVKGANDN PLLGGFTLTG FEGWGGSNDS FNTYMTGTFA ENLPTKFLQH | 900 |
| YLVYKWENYE DGQSPAGNHE KQITLKNEAG DTVVVTRKEE QREDNYIERI ITLNGKKVLE | 960 |
| DVTYLLPWTD SDTNKEKLYH YNYEGGTTTW ELQDDWADLK NVIVYKLTDT GRTEEQTVAV | 1020 |
| VDGTVTLTAE ANTPYVVVKG EEAPKTVTSW SNGAHVKDTG FNSYAGTGAG DALDENVWSG | 1080 |
| DVKNDNVKVV RVATGNKYLS MGSEEQGLEV TTAIKDLTPG KNYVAEVYVD NKSDAKAWIE | 1140 |
| VSAGTETVSN YTLKSFAGNY VQCDAHNNRG MANSNMQVML VSFTAEKAIA SLTLKREAGT | 1200 |
| GITYFDDIRI VEKTLNNIQE DGSFKQDFEE VVQGIYPFVI GSAQGVTDQV THLAEKHDPY | 1260 |
| TQSGWGNVIL DDVIGGTWSL KHHGNNSGII YQTIPQNLHL EPGVTYKVSF DYQAGRDNMY | 1320 |
| RIVTGDGDQI TQTFDYLAGT AASAVGGKST TGKFEFTITG SGTGQSWFGL SSTSFTEQKG | 1380 |
| VDYCYGQKDF ILDNLVVTPQ EGLTLNETDV ELKGVGDSRK LTASQEVTWS TSDENVVTVG | 1440 |
| TDGYIQAAGA GTATITATAK TNTALTASCT VTVATSSEVV NESPFEAVAA NTEQGGDEGK | 1500 |
| EKAIDGNTST HWHSNWSTGF TVSETNPAIL TATLKDDAKD FGGFYITQRL GNVNGLISKL | 1560 |
| TYVAGNTFDA ATNTVSDVVK EGTVEVPSAS VSAGATFGVA FGENINAKYL QIRVLNGGGN | 1620 |
| FAAIAELRTC LKVAFTDEEL LEEKQNAKTA SDAAATTADE ALTNAQTALT AAKAAEAAAE | 1680 |
| DKIAANVAVK EAELAVKQAE LNKQEAVKRQ QEAAVALALM EASQSEDATV KQQKLAAAET | 1740 |
| AKTAIATAET EILSINAALT MARYDAITAK SALEVQTKAA AVNDAKTVYD STLQPLTDAN | 1800 |
| SKLEQAKAAK EEADKTGTEL EKAEANLRLA EAELAVAEAV QNQALAQCAW ETAKAEAASA | 1860 |
| RANVAATDEE ATSAREEEKT ANEAAAAARV QIANGKKTIS ACNSSLAKAE KVIQDLKSST | 1920 |
| TELAEALDKA EAVYKAGGAA YTEASWKKFV TAYENARAAV GDSSKTPDEL YELQKTLEDA | 1980 |
| QKALEKKPDV QDSLAEAKKT LSETLKAAEA VYAAGGAAYT EASWKAFAAA YAAAKNPAAN | 2040 |
| ADAAALKALA EALKKAQSGL VKAATPAPAP TEKLKKGDTV TVKNVVYKVT NADKKTVAAV | 2100 |
| KGKNKKLASV SIASTVKVKG ETCKVTAVNA KAFKGYARLK KVTIGKNVTT VGKQAFYGCK | 2160 |
| KLTKVSFKGT AVKSIKSAAF KKANAKIKVT LPKKLKGKNR TKLVKQLKKA GIKSAK | 2216 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH101 | 324 | 1039 | 2.2e-236 | 0.9943422913719944 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12905 | Glyco_hydro_101 | 2.86e-125 | 559 | 884 | 1 | 272 | Endo-alpha-N-acetylgalactosaminidase. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by the S. pneumoniae protein Endo-alpha-N-acetylgalactosaminidase, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. |
| cd14244 | GH_101_like | 8.99e-109 | 572 | 908 | 1 | 298 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam18080 | Gal_mutarotas_3 | 2.90e-67 | 320 | 558 | 1 | 243 | Galactose mutarotase-like fold domain. This domain is found in endo-alpha-N-acetylgalactosaminidase present in Streptococcus pneumoniae. Endo-alpha-N-acetylgalactosaminidase is a cell surface-anchored glycoside hydrolase involved in the breakdown of mucin type O-linked glycans. The domain, known as domain 2, exhibits strong structural similarlity to the galactose mutarotase-like fold but lacks the active site residues. Domains, found in a number of glycoside hydrolases, structurally similar to domain 2 confer stability to the multidomain architectures. |
| pfam17974 | GalBD_like | 6.31e-64 | 1206 | 1374 | 1 | 173 | Galactose-binding domain-like. Proteins containing a galactose-binding domain-like fold can be found in several different protein families, in both eukaryotes and prokaryotes. The common function of these domains is to bind to specific ligands, such as cell-surface-attached carbohydrate substrates for galactose oxidase and sialidase, phospholipids on the outer side of the mammalian cell membrane for coagulation factor Va, membrane-anchored ephrin for the Eph family of receptor tyrosine kinases, and a complex of broken single-stranded DNA and DNA polymerase beta for XRCC1. The structure of the galactose-binding domain-like members consists of a beta-sandwich, in which the strands making up the sheets exhibit a jellyroll fold. |
| pfam17451 | Glyco_hyd_101C | 6.86e-32 | 891 | 1013 | 1 | 111 | Glycosyl hydrolase 101 beta sandwich domain. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by a S. pneumoniae protein, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. This domain represents C-terminal the beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM14139.1 | 0.0 | 42 | 1643 | 248 | 1820 |
| QWH41973.1 | 2.65e-274 | 40 | 1395 | 58 | 1445 |
| APC49531.1 | 1.90e-273 | 27 | 1395 | 24 | 1427 |
| BAL17718.1 | 3.86e-272 | 40 | 1395 | 32 | 1419 |
| AYY26584.1 | 8.14e-272 | 40 | 1395 | 58 | 1445 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2ZXQ_A | 4.42e-239 | 314 | 1628 | 21 | 1370 | Crystalstructure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) [Bifidobacterium longum] |
| 6QEP_A | 1.34e-234 | 313 | 1437 | 5 | 1157 | EngBFDARPin Fusion 4b H14 [Bifidobacterium longum] |
| 6QEV_B | 2.82e-234 | 320 | 1437 | 12 | 1157 | EngBFDARPin Fusion 4b B6 [Bifidobacterium longum] |
| 6SH9_B | 2.82e-234 | 320 | 1437 | 12 | 1157 | EngBFDARPin Fusion 4b D12 [Bifidobacterium longum subsp. longum JCM 1217] |
| 6QFK_A | 2.82e-234 | 320 | 1437 | 12 | 1157 | EngBFDARPin Fusion 4b G10 [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2MGH6 | 4.26e-206 | 58 | 1627 | 153 | 1622 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| Q8DR60 | 1.12e-205 | 58 | 1556 | 153 | 1559 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
| A9WNA0 | 1.32e-129 | 320 | 1397 | 52 | 1034 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
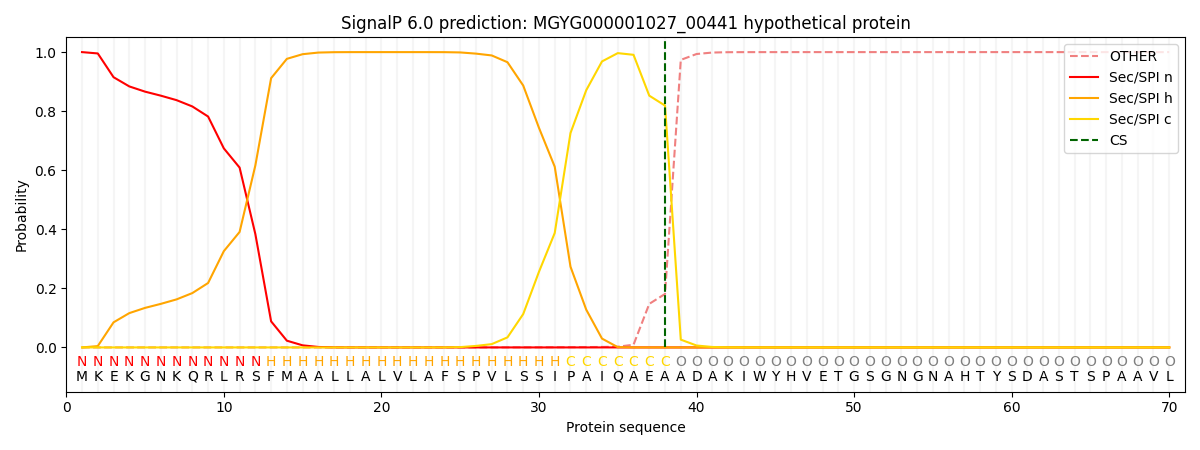
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000535 | 0.998537 | 0.000205 | 0.000288 | 0.000223 | 0.000185 |

