You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000924_00518
You are here: Home > Sequence: MGYG000000924_00518
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
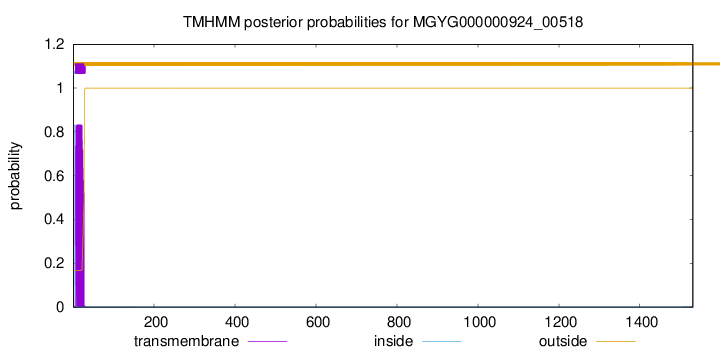
TMHMM annotations
Basic Information help
| Species | An172 sp002160515 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; An172; An172 sp002160515 | |||||||||||
| CAZyme ID | MGYG000000924_00518 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7343; End: 11938 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 249 | 538 | 8.7e-101 | 0.9966101694915255 |
| CBM32 | 1389 | 1524 | 1.1e-18 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 1.11e-121 | 249 | 536 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| cd14256 | Dockerin_I | 1.40e-13 | 110 | 165 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00754 | F5_F8_type_C | 3.36e-13 | 1389 | 1524 | 7 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02838 | Glyco_hydro_20b | 2.38e-11 | 38 | 242 | 1 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 5.33e-11 | 713 | 836 | 2 | 120 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYE33607.1 | 1.91e-285 | 39 | 1399 | 45 | 1499 |
| QAS61771.1 | 1.91e-285 | 39 | 1399 | 45 | 1499 |
| AIY84741.1 | 2.45e-247 | 1 | 1372 | 1 | 1313 |
| QMI86524.1 | 5.49e-243 | 161 | 1324 | 72 | 1239 |
| QQV04689.1 | 8.39e-242 | 25 | 1351 | 31 | 1437 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PWI_A | 1.16e-169 | 16 | 686 | 11 | 621 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 6PV4_A | 1.53e-124 | 16 | 698 | 11 | 653 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 5MI4_A | 4.60e-77 | 191 | 841 | 87 | 720 | BtGH84mutant with covalent modification by MA3 [Bacteroides thetaiotaomicron VPI-5482],5MI5_A BtGH84 mutant with covalent modification by MA3 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482],5MI6_A BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G [Bacteroides thetaiotaomicron VPI-5482],5MI7_A BtGH84 mutant with covalent modification by MA4 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482] |
| 2J4G_A | 7.32e-76 | 191 | 841 | 75 | 708 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482],2J4G_B Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482] |
| 2JIW_A | 7.48e-76 | 191 | 841 | 76 | 709 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine [Bacteroides thetaiotaomicron VPI-5482],2JIW_B Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 4.41e-205 | 1 | 1404 | 1 | 1534 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q89ZI2 | 6.24e-75 | 191 | 841 | 97 | 730 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q8XL08 | 3.44e-65 | 178 | 830 | 109 | 745 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 6.10e-64 | 178 | 830 | 109 | 745 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| O60502 | 3.07e-23 | 250 | 507 | 63 | 331 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
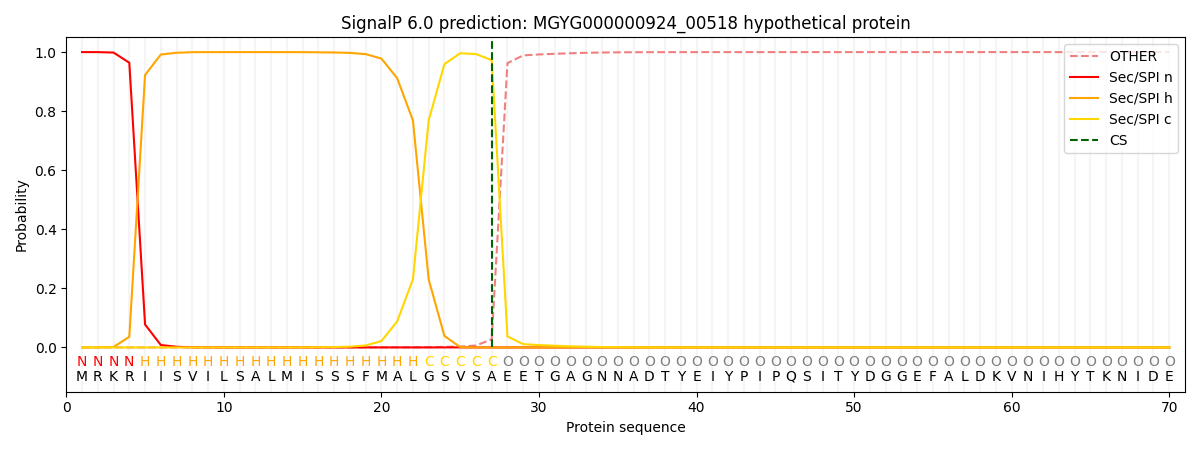
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000296 | 0.998951 | 0.000214 | 0.000198 | 0.000173 | 0.000152 |