You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000870_00023
You are here: Home > Sequence: MGYG000000870_00023
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
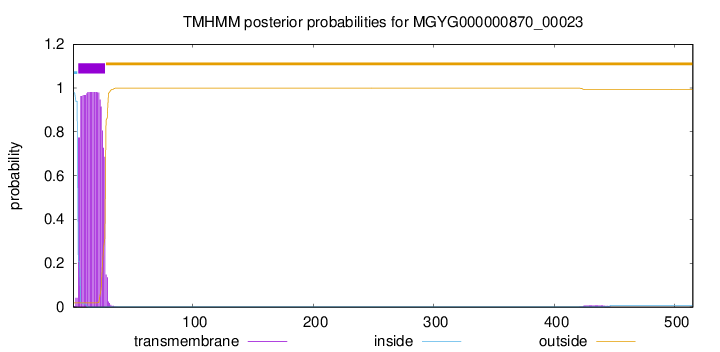
TMHMM annotations
Basic Information help
| Species | Akkermansia sp004167605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp004167605 | |||||||||||
| CAZyme ID | MGYG000000870_00023 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | Periplasmic trehalase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21048; End: 22595 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH37 | 207 | 426 | 6e-28 | 0.4378818737270876 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01204 | Trehalase | 6.57e-21 | 243 | 384 | 318 | 456 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| pfam03200 | Glyco_hydro_63 | 7.94e-19 | 170 | 428 | 195 | 487 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| COG1626 | TreA | 9.02e-19 | 207 | 392 | 325 | 510 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| PRK10137 | PRK10137 | 4.67e-18 | 250 | 382 | 598 | 738 | alpha-glucosidase; Provisional |
| PRK13272 | treA | 1.36e-10 | 260 | 426 | 366 | 525 | alpha,alpha-trehalase TreA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWO96172.1 | 0.0 | 1 | 515 | 1 | 515 |
| QWP70936.1 | 0.0 | 1 | 515 | 1 | 515 |
| QWO91044.1 | 0.0 | 1 | 515 | 1 | 515 |
| QWO93599.1 | 0.0 | 1 | 515 | 1 | 515 |
| QWO86291.1 | 0.0 | 1 | 515 | 1 | 515 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4WVA_A | 7.71e-14 | 272 | 384 | 274 | 383 | Crystalstructure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris [Thermus thermophilus HB8],4WVA_B Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris [Thermus thermophilus HB8],4WVB_A Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose [Thermus thermophilus HB8],4WVB_B Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose [Thermus thermophilus HB8],4WVC_A Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate [Thermus thermophilus HB8],4WVC_B Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate [Thermus thermophilus HB8] |
| 2Z07_A | 1.37e-13 | 272 | 384 | 274 | 383 | Crystalstructure of uncharacterized conserved protein from Thermus thermophilus HB8 [Thermus thermophilus],2Z07_B Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8 [Thermus thermophilus] |
| 5M4A_A | 5.94e-11 | 259 | 450 | 384 | 578 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae in complex with trehalose [Saccharomyces cerevisiae] |
| 5NIS_A | 6.29e-11 | 259 | 450 | 437 | 631 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae [Saccharomyces cerevisiae S288C] |
| 5JTA_A | 6.78e-11 | 259 | 450 | 532 | 726 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94250 | 3.39e-37 | 42 | 514 | 3 | 464 | Uncharacterized protein BB_0381 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0381 PE=4 SV=2 |
| O14255 | 4.12e-15 | 165 | 437 | 528 | 807 | Probable mannosyl-oligosaccharide glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC6G10.09 PE=3 SV=1 |
| P49381 | 1.21e-14 | 256 | 391 | 531 | 667 | Cytosolic neutral trehalase OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=NTH1 PE=1 SV=1 |
| O42622 | 4.55e-13 | 239 | 393 | 497 | 650 | Cytosolic neutral trehalase OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=NTH1 PE=2 SV=2 |
| O42783 | 6.06e-13 | 252 | 391 | 516 | 656 | Cytosolic neutral trehalase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=treB PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
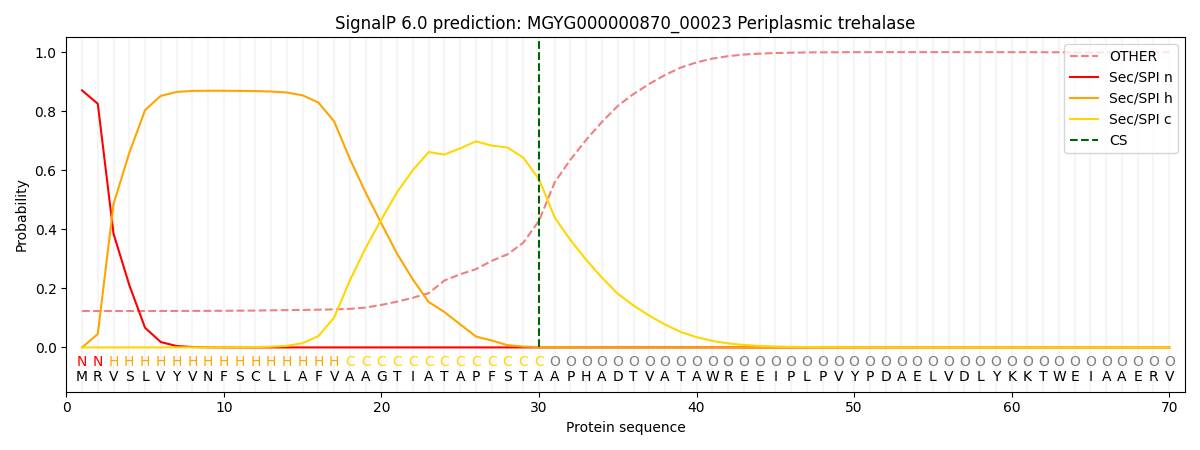
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.132938 | 0.858151 | 0.007535 | 0.000547 | 0.000353 | 0.000455 |