You are browsing environment: HUMAN GUT
MGYG000000675_01835
Basic Information
help
Species
Bacteroides congonensis
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides congonensis
CAZyme ID
MGYG000000675_01835
CAZy Family
GH30
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000675
5782172
MAG
Kazakhstan
Asia
Gene Location
Start: 24402;
End: 25910
Strand: -
No EC number prediction in MGYG000000675_01835.
Family
Start
End
Evalue
family coverage
GH30
45
497
1.7e-34
0.9688249400479616
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG5520
XynC
2.43e-11
44
454
30
413
O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis].
more
pfam11790
Glyco_hydro_cc
0.005
169
340
47
193
Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781.
more
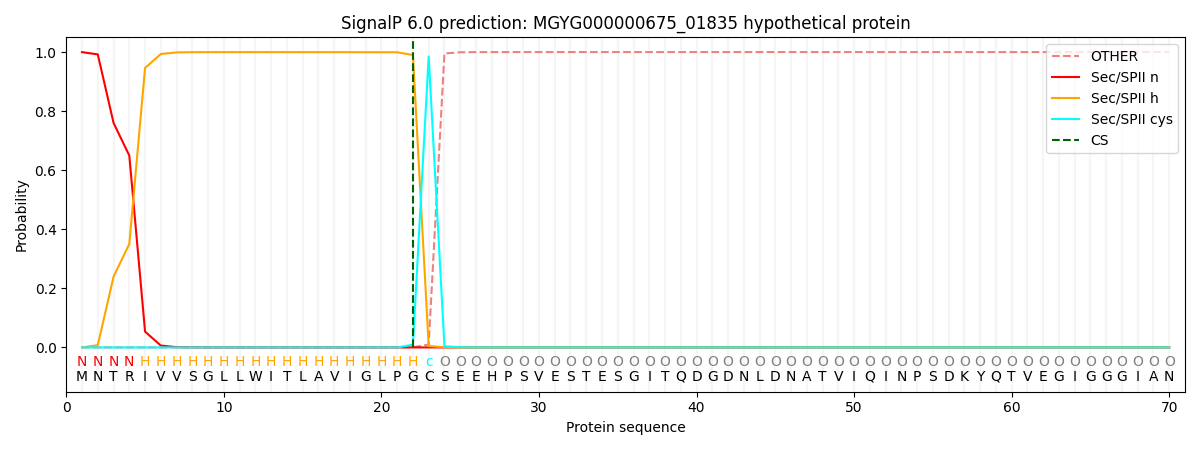
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000189
0.999876
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000000675_01835.