You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000666_01692
You are here: Home > Sequence: MGYG000000666_01692
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; ; | |||||||||||
| CAZyme ID | MGYG000000666_01692 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5542; End: 7404 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 71 | 256 | 2.6e-28 | 0.5011600928074246 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 7.57e-136 | 2 | 615 | 4 | 609 |
| QDU57372.1 | 7.51e-85 | 19 | 614 | 19 | 622 |
| AVM44901.1 | 3.16e-55 | 1 | 571 | 1 | 583 |
| AHF90941.1 | 5.72e-33 | 20 | 612 | 226 | 840 |
| ADE82856.1 | 2.29e-32 | 17 | 615 | 27 | 399 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 5.72e-10 | 23 | 251 | 101 | 345 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 1HIZ_A | 2.93e-06 | 41 | 138 | 39 | 159 | XylanaseT6 (Xt6) from Bacillus Stearothermophilus [Geobacillus stearothermophilus],1R85_A Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6): The WT enzyme (monoclinic form) at 1.45A resolution [Geobacillus stearothermophilus],1R87_A Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6, monoclinic form): The complex of the WT enzyme with xylopentaose at 1.67A resolution [Geobacillus stearothermophilus] |
| 3MMD_A | 2.93e-06 | 41 | 138 | 39 | 159 | ChainA, Endo-1,4-beta-xylanase [Geobacillus stearothermophilus] |
| 4PUD_A | 6.65e-06 | 41 | 138 | 31 | 151 | ExtracellulrXylanase from Geobacillus stearothermophilus: E159Q mutant, with xylopentaose in active site [Geobacillus stearothermophilus],4PUE_A Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylotetraose in active site [Geobacillus stearothermophilus] |
| 4PRW_A | 6.79e-06 | 41 | 138 | 39 | 159 | XylanaseT6 (XT6) from Geobacillus Stearothermophilus in complex with xylohexaose [Geobacillus stearothermophilus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
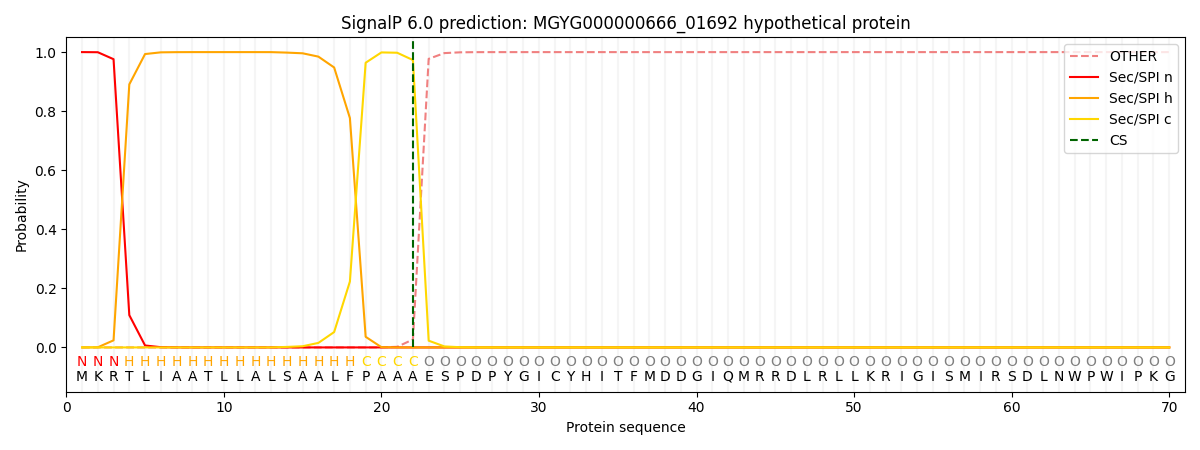
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000243 | 0.999076 | 0.000177 | 0.000173 | 0.000157 | 0.000141 |
