You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000666_00161
You are here: Home > Sequence: MGYG000000666_00161
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; ; | |||||||||||
| CAZyme ID | MGYG000000666_00161 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35563; End: 38913 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 389 | 862 | 2.9e-29 | 0.5172872340425532 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.57e-13 | 475 | 957 | 99 | 542 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00703 | Glyco_hydro_2 | 2.10e-09 | 548 | 658 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02836 | Glyco_hydro_2_C | 1.37e-05 | 667 | 889 | 8 | 212 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| TIGR04523 | Mplasa_alph_rch | 0.001 | 254 | 366 | 457 | 562 | helix-rich Mycoplasma protein. Members of this family occur strictly within a subset of Mycoplasma species. Members average 750 amino acids in length, including signal peptide. Sequences are predicted (Jpred 3) to be almost entirely alpha-helical. These sequences show strong periodicity (consistent with long alpha helical structures) and low complexity rich in D,E,N,Q, and K. Genes encoding these proteins are often found in tandem. The function is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44993.1 | 1.27e-297 | 3 | 1114 | 55 | 1225 |
| QUH31336.1 | 1.12e-19 | 391 | 890 | 1 | 467 |
| AIQ32428.1 | 1.17e-17 | 391 | 890 | 1 | 518 |
| AIQ71089.1 | 2.59e-17 | 528 | 1028 | 171 | 657 |
| QSF44046.1 | 2.62e-17 | 391 | 1019 | 1 | 674 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T98_A | 1.36e-12 | 482 | 886 | 108 | 499 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 7CWD_A | 6.34e-09 | 462 | 791 | 69 | 405 | ChainA, beta-glalactosidase [Niallia circulans],7CWI_A Chain A, beta-galactosidase [Niallia circulans] |
| 4CU6_A | 2.52e-08 | 463 | 791 | 76 | 429 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU7_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU8_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4] |
| 4CUC_A | 2.52e-08 | 463 | 791 | 76 | 429 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA. [Streptococcus pneumoniae TIGR4] |
| 3DEC_A | 4.63e-06 | 443 | 790 | 100 | 460 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
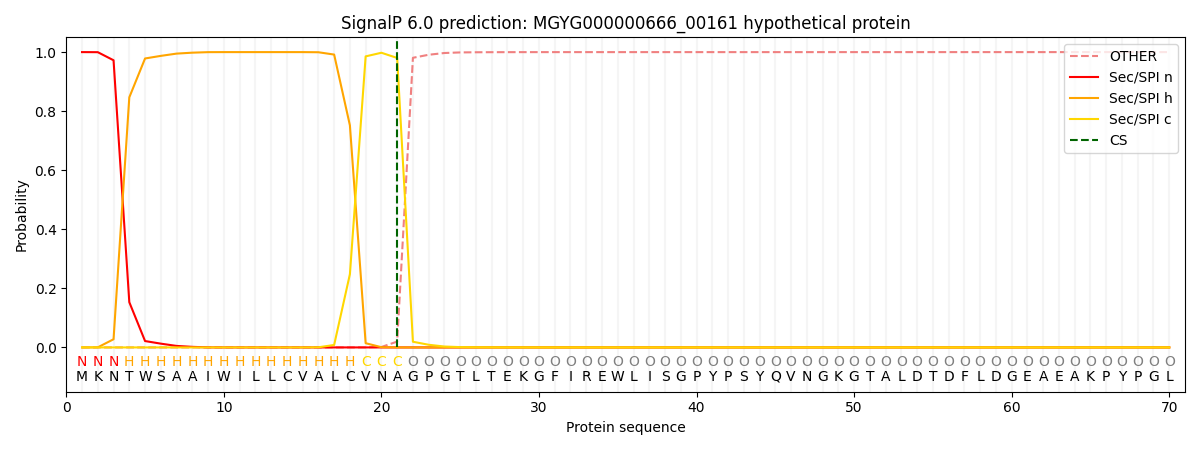
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000184 | 0.999224 | 0.000151 | 0.000148 | 0.000140 | 0.000131 |

