You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000526_02343
You are here: Home > Sequence: MGYG000000526_02343
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900770395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900770395 | |||||||||||
| CAZyme ID | MGYG000000526_02343 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 69074; End: 71599 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 502 | 823 | 8e-80 | 0.9888475836431226 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07632 | DUF1593 | 6.80e-99 | 29 | 312 | 1 | 261 | Protein of unknown function (DUF1593). A family of proteins in Rhodopirellula baltica that are predicted to be secreted. Also, a member has been identified in Caulobacter crescentus. These proteins mat be related to pfam01156. |
| pfam18911 | PKD_4 | 3.30e-05 | 361 | 406 | 9 | 48 | PKD domain. This entry is composed of PKD domains found in bacterial surface proteins. |
| pfam02010 | REJ | 0.002 | 385 | 430 | 100 | 155 | REJ domain. The REJ (Receptor for Egg Jelly) domain is found in PKD1 and the sperm receptor for egg jelly. The function of this domain is unknown. The domain is 600 amino acids long so is probably composed of multiple structural domains. There are six completely conserved cysteine residues that may form disulphide bridges. This region contains tandem PKD-like domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| EFV29552.1 | 9.75e-185 | 473 | 839 | 28 | 395 |
| QUT46099.1 | 1.38e-184 | 473 | 839 | 28 | 395 |
| QRQ48279.1 | 1.38e-184 | 473 | 839 | 28 | 395 |
| ADD61496.1 | 1.04e-177 | 473 | 839 | 28 | 395 |
| QUT89422.1 | 1.53e-177 | 473 | 840 | 29 | 397 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YHG_A | 8.76e-69 | 29 | 463 | 30 | 435 | Abinitio phasing of a nucleoside hydrolase-related hypothetical protein from Saccharophagus degradans that is associated with carbohydrate metabolism [Saccharophagus degradans 2-40] |
| 4G4G_A | 6.84e-64 | 502 | 834 | 59 | 399 | Crystalstructure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution [Thermothelomyces thermophilus ATCC 42464] |
| 4G4I_A | 1.80e-63 | 502 | 834 | 59 | 399 | Crystalstructure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile determined at 1.9 A resolution [Thermothelomyces thermophilus ATCC 42464],4G4J_A Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution [Thermothelomyces thermophilus ATCC 42464] |
| 3PIC_A | 2.49e-61 | 502 | 836 | 26 | 358 | ChainA, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
| 6RU2_A | 1.81e-59 | 502 | 832 | 18 | 354 | CrystalStructure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2QJR6 | 1.05e-63 | 460 | 834 | 9 | 385 | 4-O-methyl-glucuronoyl methylesterase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=ge2 PE=1 SV=1 |
| Q7S1X0 | 3.41e-62 | 482 | 823 | 16 | 364 | 4-O-methyl-glucuronoyl methylesterase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=Cip2 PE=1 SV=2 |
| B2ABS0 | 8.54e-62 | 502 | 835 | 123 | 460 | 4-O-methyl-glucuronoyl methylesterase OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=ge1 PE=1 SV=1 |
| D8QLP9 | 4.22e-60 | 502 | 831 | 36 | 366 | 4-O-methyl-glucuronoyl methylesterase OS=Schizophyllum commune (strain H4-8 / FGSC 9210) OX=578458 GN=SCHCODRAFT_238770 PE=1 SV=1 |
| G0RV93 | 1.21e-59 | 502 | 836 | 111 | 443 | 4-O-methyl-glucuronoyl methylesterase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cip2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
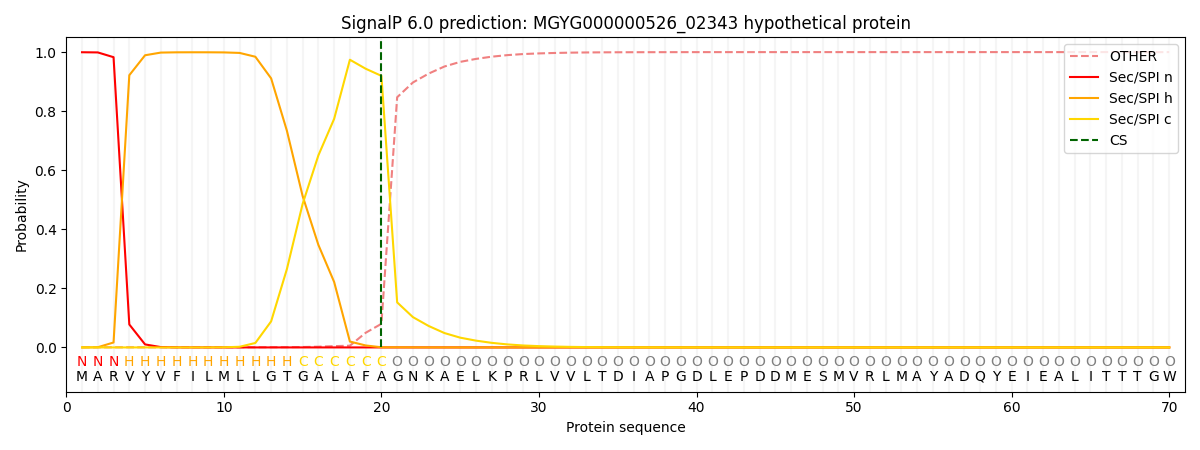
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001605 | 0.997439 | 0.000311 | 0.000217 | 0.000203 | 0.000198 |
