You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000484_00575
You are here: Home > Sequence: MGYG000000484_00575
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
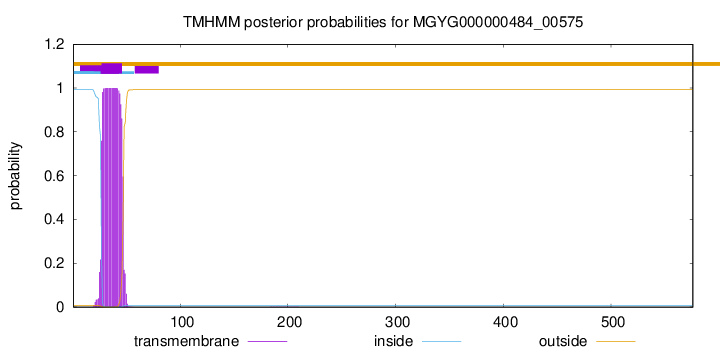
TMHMM annotations
Basic Information help
| Species | Butyrivibrio_A sp000431815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Butyrivibrio_A; Butyrivibrio_A sp000431815 | |||||||||||
| CAZyme ID | MGYG000000484_00575 | |||||||||||
| CAZy Family | CBM2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 249636; End: 251366 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 293 | 533 | 2.1e-96 | 0.9915611814345991 |
| CBM2 | 116 | 214 | 9.5e-19 | 0.9504950495049505 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.96e-72 | 291 | 540 | 1 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 8.27e-18 | 275 | 479 | 37 | 227 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| smart00637 | CBD_II | 3.72e-13 | 121 | 215 | 1 | 92 | CBD_II domain. |
| pfam00553 | CBM_2 | 2.22e-12 | 115 | 215 | 2 | 100 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart01063 | CBM49 | 9.40e-05 | 119 | 192 | 5 | 74 | Carbohydrate binding domain CBM49. This domain is found at the C terminal of cellulases and in vitro binding studies have shown it to binds to crystalline cellulose. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT53734.1 | 1.40e-146 | 122 | 576 | 81 | 515 |
| QNM00780.1 | 4.47e-145 | 122 | 576 | 81 | 515 |
| CBK83877.1 | 9.19e-140 | 123 | 576 | 87 | 530 |
| ADD61840.1 | 1.59e-128 | 109 | 576 | 65 | 539 |
| ACR73731.1 | 4.57e-127 | 109 | 576 | 65 | 536 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 5.10e-110 | 275 | 573 | 5 | 300 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZW_A | 2.26e-102 | 275 | 570 | 4 | 299 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 3PZT_A | 4.47e-100 | 275 | 575 | 29 | 326 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZB_A | 2.30e-99 | 275 | 570 | 4 | 300 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 1H11_A | 1.34e-91 | 272 | 573 | 1 | 300 | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYMEINTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION [Salipaludibacillus agaradhaerens],1H2J_A ENDOGLUCANASE CEL5A IN COMPLEX WITH UNHYDROLYSED AND COVALENTLY LINKED 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-CELLOBIOSIDE AT 1.15 A RESOLUTION [Salipaludibacillus agaradhaerens],1HF6_A ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE [Salipaludibacillus agaradhaerens],1OCQ_A COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine [Salipaludibacillus agaradhaerens],1W3K_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellobio Derived-tetrahydrooxazine [Salipaludibacillus agaradhaerens],1W3L_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellotri Derived-Tetrahydrooxazine [Salipaludibacillus agaradhaerens],4A3H_A 2',4' Dinitrophenyl-2-Deoxy-2-Fluro-B-D-Cellobioside Complex Of The Endoglucanase Cel5a From Bacillus Agaradhaerens At 1.6 A Resolution [Salipaludibacillus agaradhaerens],5A3H_A 2-Deoxy-2-Fluro-B-D-CellobiosylENZYME INTERMEDIATE COMPLEX Of The Endoglucanase Cel5a From Bacillus Agaradhearans At 1.8 Angstroms Resolution [Salipaludibacillus agaradhaerens],6A3H_A 2-Deoxy-2-Fluro-B-D-CellotriosylENZYME INTERMEDIATE COMPLEX OF THE Endoglucanase Cel5a From Bacillus Agaradhearans At 1.6 Angstrom Resolution [Salipaludibacillus agaradhaerens],7A3H_A Native Endoglucanase Cel5a Catalytic Core Domain At 0.95 Angstroms Resolution [Salipaludibacillus agaradhaerens],8A3H_A Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15704 | 7.07e-99 | 281 | 575 | 45 | 337 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P10475 | 4.91e-97 | 275 | 575 | 34 | 331 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 5.38e-96 | 275 | 575 | 34 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| Q59394 | 9.24e-95 | 273 | 573 | 31 | 328 | Endoglucanase N OS=Pectobacterium atrosepticum OX=29471 GN=celN PE=3 SV=1 |
| Q47096 | 2.74e-94 | 273 | 573 | 31 | 328 | Endoglucanase 5 OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=celV PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
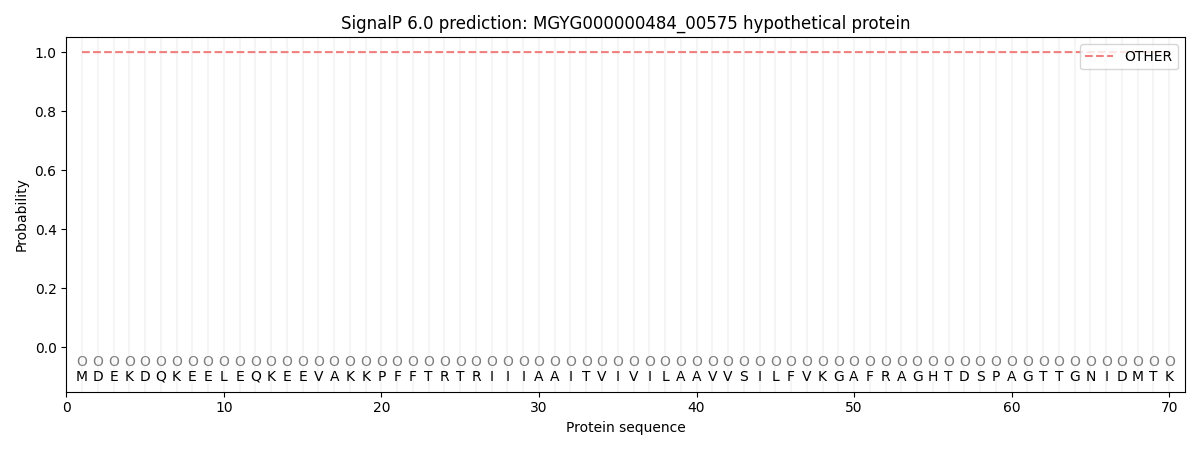
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000017 | 0.000017 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |