You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000243_00578
You are here: Home > Sequence: MGYG000000243_00578
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola massiliensis | |||||||||||
| CAZyme ID | MGYG000000243_00578 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 78969; End: 81932 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 62 | 859 | 1.3e-115 | 0.9561170212765957 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10340 | ebgA | 3.81e-71 | 69 | 795 | 44 | 808 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 2.34e-65 | 55 | 564 | 1 | 507 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK09525 | lacZ | 9.40e-58 | 51 | 446 | 30 | 463 | beta-galactosidase. |
| PRK10150 | PRK10150 | 5.60e-48 | 66 | 554 | 12 | 553 | beta-D-glucuronidase; Provisional |
| pfam02836 | Glyco_hydro_2_C | 2.38e-24 | 324 | 446 | 1 | 136 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50207.1 | 3.39e-180 | 52 | 959 | 198 | 1112 |
| QRO24898.1 | 8.00e-168 | 52 | 958 | 193 | 1098 |
| AQT68269.1 | 4.21e-164 | 53 | 985 | 265 | 1212 |
| BBE16084.1 | 1.18e-160 | 53 | 959 | 40 | 961 |
| QMV71036.1 | 1.19e-148 | 69 | 987 | 44 | 971 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 9.80e-62 | 68 | 893 | 40 | 891 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 9.85e-62 | 68 | 893 | 41 | 892 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3BGA_A | 9.38e-48 | 68 | 928 | 50 | 931 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3DEC_A | 4.26e-44 | 68 | 887 | 46 | 900 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3DYM_A | 3.21e-43 | 46 | 725 | 37 | 765 | ChainA, Beta-galactosidase [Escherichia coli K-12],3DYM_B Chain B, Beta-galactosidase [Escherichia coli K-12],3DYM_C Chain C, Beta-galactosidase [Escherichia coli K-12],3DYM_D Chain D, Beta-galactosidase [Escherichia coli K-12],3E1F_1 Chain 1, Beta-galactosidase [Escherichia coli K-12],3E1F_2 Chain 2, Beta-galactosidase [Escherichia coli K-12],3E1F_3 Chain 3, Beta-galactosidase [Escherichia coli K-12],3E1F_4 Chain 4, Beta-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 5.39e-61 | 68 | 893 | 41 | 892 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| Q9K9C6 | 1.86e-51 | 67 | 741 | 46 | 771 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| P24131 | 1.34e-50 | 116 | 877 | 124 | 897 | Beta-galactosidase OS=Clostridium acetobutylicum OX=1488 GN=cbgA PE=2 SV=2 |
| Q04F24 | 3.35e-49 | 57 | 859 | 34 | 910 | Beta-galactosidase OS=Oenococcus oeni (strain ATCC BAA-331 / PSU-1) OX=203123 GN=lacZ PE=3 SV=1 |
| A8AKB8 | 5.80e-49 | 68 | 725 | 54 | 769 | Beta-galactosidase OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
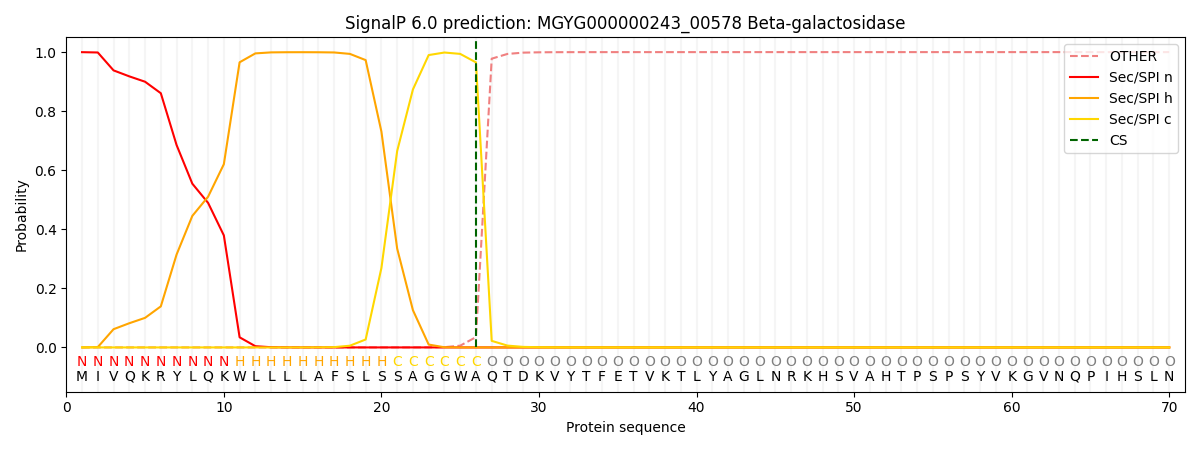
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000630 | 0.998211 | 0.000635 | 0.000177 | 0.000161 | 0.000151 |
