You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000221_01808
You are here: Home > Sequence: MGYG000000221_01808
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000221_01808 | |||||||||||
| CAZy Family | PL21 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 209489; End: 212236 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL21 | 546 | 613 | 3.5e-29 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18675 | HepII_C | 2.90e-07 | 830 | 906 | 10 | 87 | Heparinase II C-terminal domain. Heparinase II (HepII) is an 85-kDa dimeric enzyme that depolymerizes both heparin and heparan sulfate glycosaminoglycans. The protein is composed of three domains: an N-terminal alpha-helical domain, a central two-layered beta-sheet domain, and a C-terminal domain forming a two-layered beta-sheet. The C-terminal domain contains nine beta-strands packed together in a manner resembling a beta-barrel. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCD19147.1 | 0.0 | 5 | 912 | 6 | 915 |
| QJB33697.1 | 1.85e-312 | 47 | 909 | 32 | 896 |
| QJB40222.1 | 5.26e-312 | 47 | 909 | 32 | 896 |
| QQS98221.1 | 2.78e-310 | 89 | 911 | 67 | 900 |
| QQT26180.1 | 6.55e-309 | 89 | 911 | 88 | 921 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2FUQ_A | 3.82e-96 | 197 | 903 | 19 | 743 | ChainA, heparinase II protein [Pedobacter heparinus],2FUQ_B Chain B, heparinase II protein [Pedobacter heparinus] |
| 3E80_A | 3.99e-96 | 197 | 903 | 21 | 745 | Structureof Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus],3E80_B Structure of Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus],3E80_C Structure of Heparinase II complexed with heparan sulfate degradation disaccharide product [Pedobacter heparinus] |
| 3E7J_A | 2.07e-93 | 197 | 903 | 21 | 745 | ChainA, Heparinase II protein [Pedobacter heparinus],3E7J_B Chain B, Heparinase II protein [Pedobacter heparinus] |
| 2FUT_A | 1.45e-92 | 197 | 903 | 20 | 744 | ChainA, heparinase II protein [Pedobacter heparinus],2FUT_B Chain B, heparinase II protein [Pedobacter heparinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6XZB6 | 3.67e-95 | 197 | 903 | 44 | 768 | Heparin and heparin-sulfate lyase OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / CIP 104194 / JCM 7457 / NBRC 12017 / NCIMB 9290 / NRRL B-14731 / HIM 762-3) OX=485917 GN=hepB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
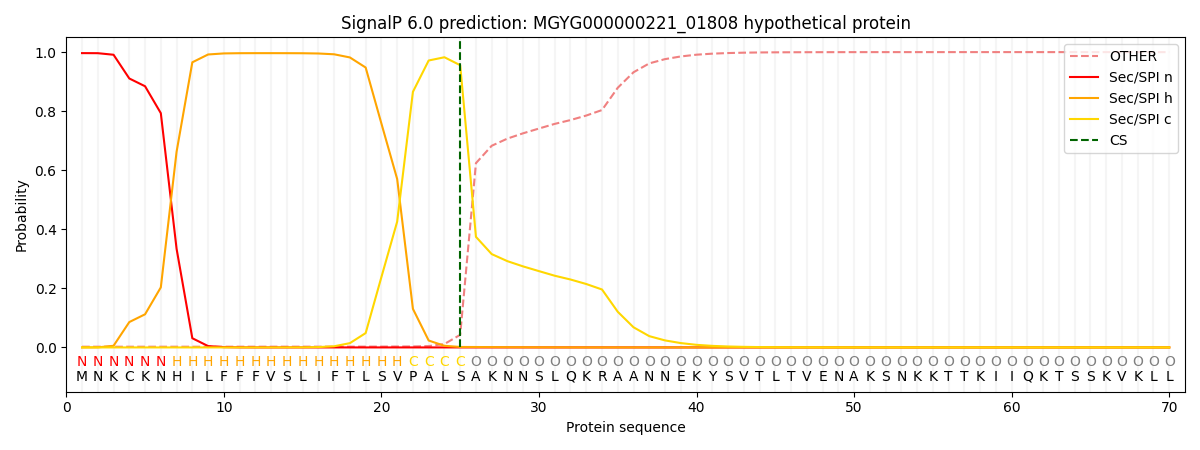
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003698 | 0.994381 | 0.001307 | 0.000211 | 0.000184 | 0.000178 |

