You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000169_00207
You are here: Home > Sequence: MGYG000000169_00207
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
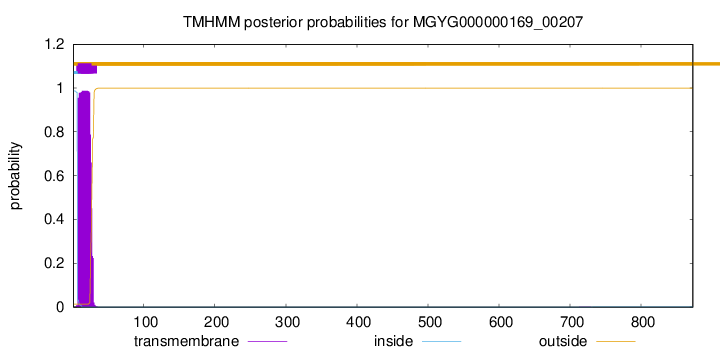
TMHMM annotations
Basic Information help
| Species | Eubacterium_G sp000432355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_G; Eubacterium_G sp000432355 | |||||||||||
| CAZyme ID | MGYG000000169_00207 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 234541; End: 237162 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 387 | 685 | 1.4e-62 | 0.4783783783783784 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17481 | Phage_sheath_1N | 0.006 | 721 | 785 | 4 | 78 | Phage tail sheath protein beta-sandwich domain. This entry represents the N-terminal beta sandwich domain found in a variety of phage tail sheath proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADQ45037.1 | 3.58e-181 | 38 | 697 | 1373 | 2007 |
| ADQ45027.1 | 2.82e-179 | 38 | 728 | 1388 | 2049 |
| QUC58319.1 | 6.18e-174 | 119 | 695 | 189 | 727 |
| ALV31537.1 | 1.33e-172 | 119 | 695 | 188 | 726 |
| AVK46774.1 | 2.23e-172 | 44 | 697 | 87 | 754 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4TZ1_A | 6.63e-139 | 119 | 680 | 9 | 532 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4TZ3_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4TZ5_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4TZ5_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4TYV_A | 7.07e-139 | 119 | 680 | 11 | 534 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4TYV_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E] |
| 4PEX_A | 9.69e-139 | 119 | 680 | 21 | 544 | Structureof the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEX_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEY_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4PEZ_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4PF0_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4PF0_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4PEW_A | 1.50e-137 | 119 | 680 | 21 | 544 | Structureof sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4PEW_B Structure of sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2NFJ9 | 2.68e-138 | 119 | 680 | 65 | 588 | Exo-beta-1,3-glucanase OS=Streptomyces sp. (strain SirexAA-E / ActE) OX=862751 GN=SACTE_4363 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000258 | 0.999069 | 0.000175 | 0.000187 | 0.000150 | 0.000137 |