You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000164_02193
You are here: Home > Sequence: MGYG000000164_02193
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
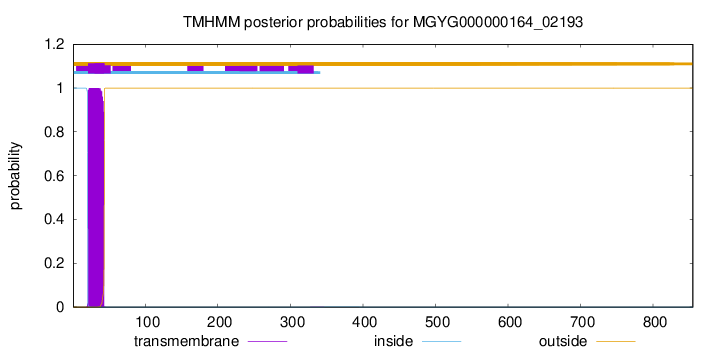
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter_A butyricigenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter_A; Mediterraneibacter_A butyricigenes | |||||||||||
| CAZyme ID | MGYG000000164_02193 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 221915; End: 224482 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 318 | 435 | 2.5e-21 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 1.03e-20 | 707 | 840 | 26 | 160 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG4193 | LytD | 1.64e-19 | 266 | 435 | 50 | 228 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG5492 | YjdB | 2.22e-19 | 549 | 790 | 20 | 306 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam07538 | ChW | 4.26e-17 | 758 | 792 | 1 | 35 | Clostridial hydrophobic W. A novel extracellular macromolecular system has been proposed based on the proteins containing ChW repeats. ChW stands for Clostridial hydrophobic with conserved W (tryptophan). This repeat was originally described in Clostridium acetobutylicum but is also found in other Gram-positive bacteria including Enterococcus faecalis, Streptococcus agalactiae and Streptomyces coelicolor. |
| pfam07538 | ChW | 4.77e-14 | 560 | 594 | 1 | 35 | Clostridial hydrophobic W. A novel extracellular macromolecular system has been proposed based on the proteins containing ChW repeats. ChW stands for Clostridial hydrophobic with conserved W (tryptophan). This repeat was originally described in Clostridium acetobutylicum but is also found in other Gram-positive bacteria including Enterococcus faecalis, Streptococcus agalactiae and Streptomyces coelicolor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK17141.1 | 3.49e-133 | 130 | 536 | 89 | 494 |
| AWY97555.1 | 1.24e-118 | 129 | 533 | 82 | 484 |
| QOV20091.1 | 5.16e-113 | 128 | 533 | 73 | 476 |
| QPS13527.1 | 3.39e-107 | 130 | 545 | 108 | 517 |
| QMW73215.1 | 3.39e-107 | 130 | 545 | 108 | 517 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.42e-25 | 175 | 436 | 56 | 283 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 2.13e-24 | 175 | 436 | 424 | 651 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 2.47e-24 | 175 | 436 | 468 | 695 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005667 | 0.993341 | 0.000334 | 0.000241 | 0.000198 | 0.000194 |