You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000135_02335
You are here: Home > Sequence: MGYG000000135_02335
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Murimonas intestini | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Murimonas; Murimonas intestini | |||||||||||
| CAZyme ID | MGYG000000135_02335 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 312916; End: 315378 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 183 | 383 | 8.3e-27 | 0.9212962962962963 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 7.34e-35 | 130 | 540 | 1 | 369 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 6.54e-26 | 133 | 450 | 3 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 3.89e-13 | 303 | 646 | 233 | 593 | beta-glucosidase BglX. |
| pfam01915 | Glyco_hydro_3_C | 6.41e-08 | 510 | 646 | 1 | 183 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK05337 | PRK05337 | 0.002 | 219 | 359 | 91 | 249 | beta-hexosaminidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTE67446.1 | 0.0 | 59 | 820 | 29 | 795 |
| QTE67443.1 | 4.96e-139 | 57 | 818 | 23 | 837 |
| BBH86235.1 | 2.17e-38 | 96 | 563 | 10 | 482 |
| QRK06309.1 | 3.68e-38 | 93 | 563 | 29 | 517 |
| AYA66408.1 | 6.26e-38 | 92 | 538 | 36 | 507 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6JGQ_A | 2.25e-20 | 111 | 553 | 8 | 447 | Crystalstructure of barley exohydrolaseI W434Y mutant in complex with methyl 2-thio-beta-sophoroside. [Hordeum vulgare subsp. vulgare],6JGR_A Crystal structure of barley exohydrolaseI W434Y in complex with 4'-nitrophenyl thiolaminaribioside [Hordeum vulgare subsp. vulgare],6JGS_A Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. [Hordeum vulgare subsp. vulgare],6JGT_A Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 6-thio-beta-gentiobioside. [Hordeum vulgare subsp. vulgare] |
| 3WLQ_A | 2.98e-20 | 111 | 553 | 8 | 447 | CrystalStructure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],3WLR_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],6MI1_A CRYSTAL STRUCTURE ANALYSIS OF THE VARIANT PLANT EXOHYDROLASE ARG158ALA-GLU161ALA IN COMPLEX WITH METHYL 6-THIO-BETA-GENTIOBIOSIDE [Hordeum vulgare subsp. vulgare] |
| 6JGG_A | 6.90e-20 | 111 | 553 | 8 | 447 | Crystalstructure of barley exohydrolaseI W434F mutant in complex with methyl 2-thio-beta-sophoroside. [Hordeum vulgare subsp. vulgare],6JGK_A Crystal structure of barley exohydrolaseI W434F mutant in complex with 4I,4III,4V-S-trithiocellohexaose [Hordeum vulgare subsp. vulgare],6LBV_A Crystal structure of barley exohydrolaseI W434F mutant in complex with methyl 6-thio-beta-gentiobioside [Hordeum vulgare subsp. vulgare],6LC5_A Crystal structure of barley exohydrolaseI W434F in complex with 4'-nitrophenyl thiolaminaribioside [Hordeum vulgare subsp. vulgare] |
| 1LQ2_A | 8.93e-20 | 111 | 553 | 4 | 443 | Crystalstructure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X38_A crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X39_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare] |
| 1EX1_A | 9.01e-20 | 111 | 553 | 4 | 443 | Beta-d-glucanExohydrolase From Barley [Hordeum vulgare],1IEQ_A Crystal Structure Of Barley Beta-D-Glucan Glucohydrolase Isoenzyme Exo1 [Hordeum vulgare],1IEV_A Crystal Structure Of Barley Beta-d-glucan Glucohydrolase Isoenzyme Exo1 In Complex With Cyclohexitol [Hordeum vulgare],1IEW_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 2-deoxy-2-fluoro-alpha-D-glucoside [Hordeum vulgare],1IEX_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4I,4III,4V-S-trithiocellohexaose [Hordeum vulgare],1J8V_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4'-nitrophenyl 3I-thiolaminaritrioside [Hordeum vulgare],3WLH_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],3WLJ_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 3-deoxy-glucose [Hordeum vulgare subsp. vulgare],3WLK_X Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 4-deoxy-glucose [Hordeum vulgare subsp. vulgare],3WLL_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with PEG400 [Hordeum vulgare subsp. vulgare],3WLM_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with octyl-O-glucoside [Hordeum vulgare subsp. vulgare],3WLN_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with octyl-S-glucoside [Hordeum vulgare subsp. vulgare],3WLP_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 1.38e-31 | 91 | 561 | 37 | 524 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q2UFP8 | 1.08e-25 | 105 | 560 | 39 | 499 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| Q5BCC6 | 4.03e-25 | 109 | 687 | 35 | 618 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| B8NGU6 | 4.34e-25 | 111 | 560 | 41 | 495 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| A7LXU3 | 3.90e-13 | 153 | 568 | 119 | 483 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
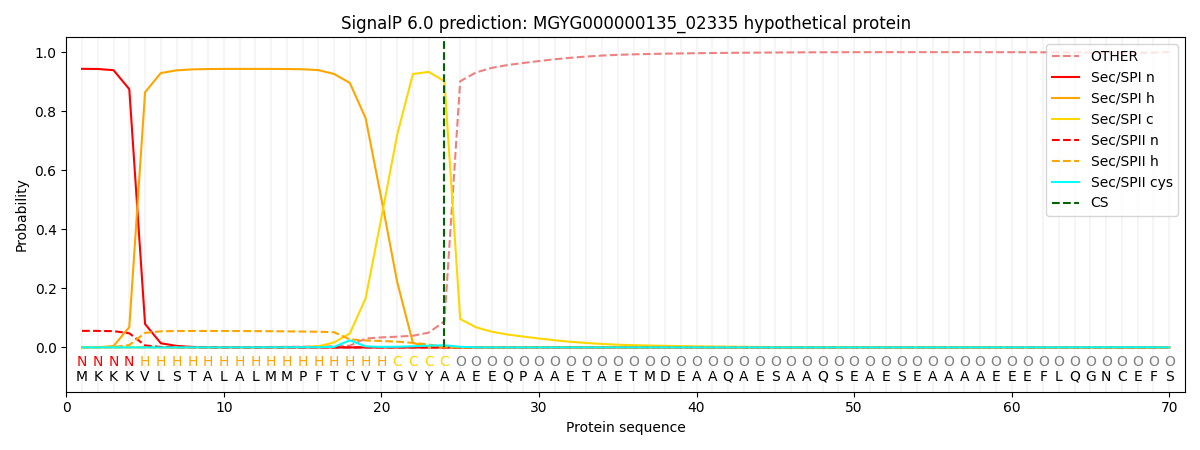
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002917 | 0.936004 | 0.059549 | 0.000908 | 0.000324 | 0.000262 |
