You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000134_01118
You are here: Home > Sequence: MGYG000000134_01118
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
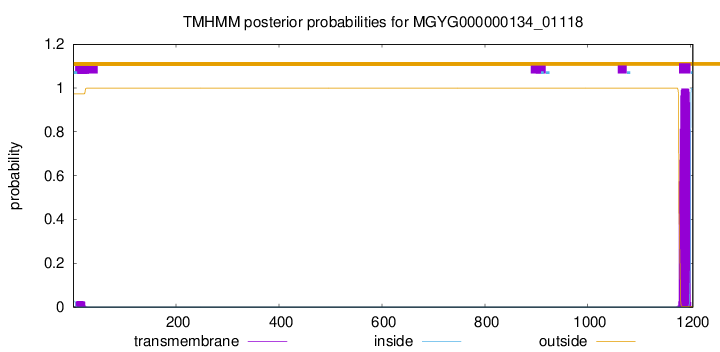
TMHMM annotations
Basic Information help
| Species | Longibaculum muris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Longibaculum; Longibaculum muris | |||||||||||
| CAZyme ID | MGYG000000134_01118 | |||||||||||
| CAZy Family | CBM4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 159223; End: 162837 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM4 | 28 | 152 | 3.2e-22 | 0.9841269841269841 |
| CBM4 | 320 | 444 | 9.2e-20 | 0.8809523809523809 |
| GH30 | 610 | 894 | 3.9e-16 | 0.592326139088729 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02018 | CBM_4_9 | 1.84e-09 | 320 | 462 | 3 | 133 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| COG4932 | COG4932 | 4.00e-08 | 958 | 1086 | 1065 | 1197 | Uncharacterized surface anchored protein [Function unknown]. |
| COG4932 | COG4932 | 6.37e-08 | 953 | 1086 | 971 | 1104 | Uncharacterized surface anchored protein [Function unknown]. |
| pfam02018 | CBM_4_9 | 6.50e-08 | 28 | 139 | 2 | 116 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| COG4932 | COG4932 | 1.25e-07 | 953 | 1086 | 1158 | 1290 | Uncharacterized surface anchored protein [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQV06119.1 | 8.88e-298 | 2 | 955 | 6 | 961 |
| QMW75430.1 | 3.55e-297 | 2 | 955 | 6 | 961 |
| QPS14235.1 | 3.55e-297 | 2 | 955 | 6 | 961 |
| QQY26901.1 | 1.00e-296 | 2 | 955 | 6 | 961 |
| QMW75427.1 | 5.77e-71 | 493 | 978 | 37 | 599 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
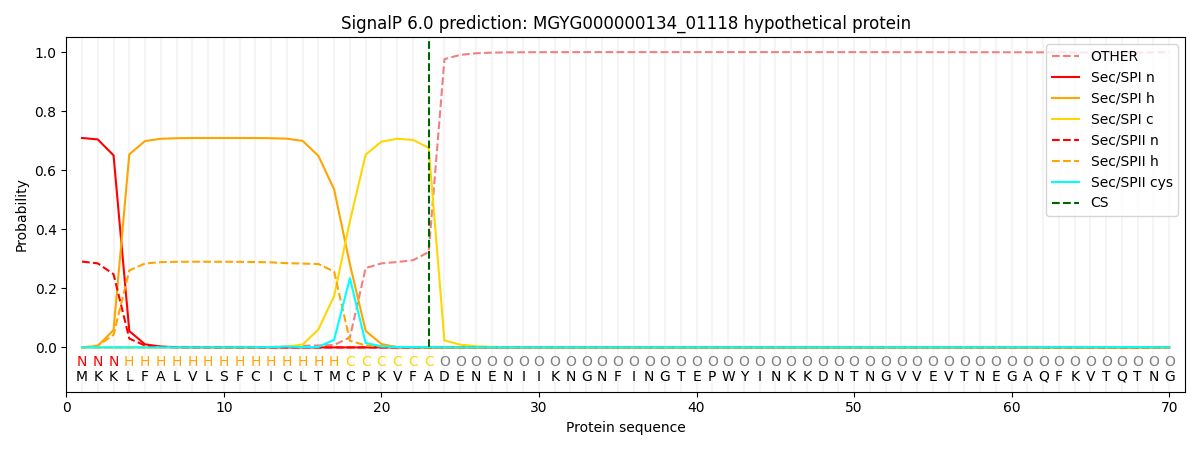
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000502 | 0.700556 | 0.298239 | 0.000234 | 0.000234 | 0.000218 |