You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000134_00881
You are here: Home > Sequence: MGYG000000134_00881
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
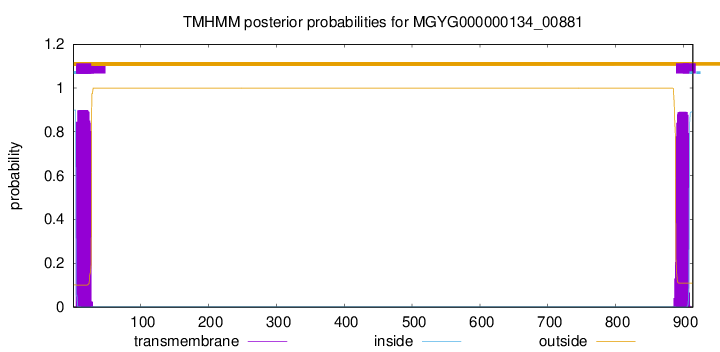
TMHMM annotations
Basic Information help
| Species | Longibaculum muris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Longibaculum; Longibaculum muris | |||||||||||
| CAZyme ID | MGYG000000134_00881 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 124552; End: 127296 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 231 | 442 | 2.3e-42 | 0.8101851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 9.77e-42 | 150 | 611 | 7 | 391 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 1.84e-27 | 174 | 515 | 27 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 2.29e-20 | 244 | 793 | 121 | 600 | beta-glucosidase BglX. |
| pfam00746 | Gram_pos_anchor | 5.91e-04 | 872 | 914 | 1 | 42 | LPXTG cell wall anchor motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QPS11706.1 | 0.0 | 20 | 913 | 17 | 893 |
| QMW75023.1 | 0.0 | 20 | 913 | 17 | 893 |
| QQV06598.1 | 0.0 | 20 | 913 | 17 | 893 |
| QQY26495.1 | 0.0 | 20 | 913 | 17 | 893 |
| AQR97785.1 | 3.13e-274 | 54 | 841 | 48 | 799 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 9.83e-18 | 242 | 796 | 142 | 633 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 6R5I_A | 8.32e-17 | 228 | 830 | 75 | 617 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 5XXL_A | 1.13e-16 | 243 | 799 | 100 | 592 | Crystalstructure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXL_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXM_A Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482],5XXM_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482] |
| 5TF0_A | 1.48e-16 | 243 | 799 | 99 | 591 | CrystalStructure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393],5TF0_B Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393] |
| 6R5O_A | 3.29e-16 | 228 | 830 | 75 | 617 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5O_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5R_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 6.84e-63 | 46 | 809 | 3 | 640 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| B8NGU6 | 7.08e-54 | 198 | 809 | 121 | 608 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q5BCC6 | 4.66e-53 | 198 | 809 | 113 | 601 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| Q2UFP8 | 6.43e-53 | 198 | 809 | 125 | 612 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| P33363 | 4.06e-20 | 243 | 830 | 120 | 649 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000284 | 0.998993 | 0.000170 | 0.000197 | 0.000182 | 0.000152 |