You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000131_02912
You are here: Home > Sequence: MGYG000000131_02912
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
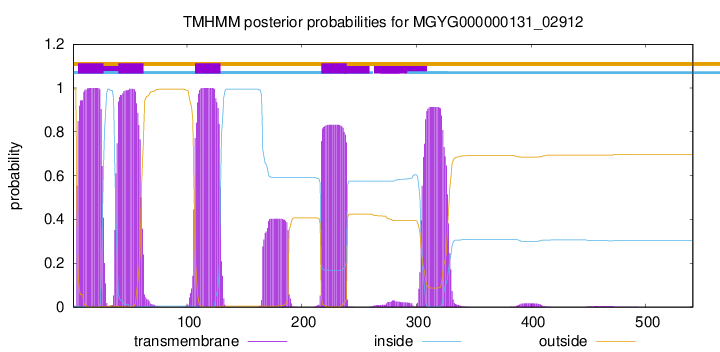
TMHMM annotations
Basic Information help
| Species | Anaerotruncus rubiinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Anaerotruncus; Anaerotruncus rubiinfantis | |||||||||||
| CAZyme ID | MGYG000000131_02912 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 75993; End: 77618 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 349 | 517 | 8.3e-57 | 0.943502824858757 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00912 | Transgly | 1.11e-70 | 348 | 517 | 9 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| TIGR02074 | PBP_1a_fam | 8.92e-66 | 353 | 537 | 3 | 186 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 2.42e-62 | 347 | 537 | 71 | 261 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 1.78e-51 | 355 | 537 | 70 | 251 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| cd07341 | M56_BlaR1_MecR1_like | 1.35e-42 | 120 | 300 | 8 | 187 | Peptidase M56-like including those in BlaR1 and MecR1, integral membrane metallopeptidase. This family contains peptidase M56, which includes zinc metalloprotease domain in MecR1 as well as BlaR1. MecR1 is a transmembrane beta-lactam sensor/signal transducer protein that regulates the expression of an altered penicillin-binding protein PBP2a, which resists inactivation by beta-lactam antibiotics, in methicillin-resistant Staphylococcus aureus (MRSA). BlaR1 regulates the inducible expression of a class A beta-lactamase that hydrolytically destroys certain ?-lactam antibiotics in MRSA. Both, MecR1 and BlaR1, are transmembrane proteins that consist of four transmembrane helices, a cytoplasmic zinc protease domain, and the soluble C-terminal extracellular sensor domain, and are highly similar in sequence and function. The signal for protein expression is transmitted by site-specific proteolytic cleavage of both the transducer, which auto-activates, and the repressor, which is inactivated, unblocking gene transcription. All members contain the zinc metalloprotease motif (HEXXH). Homologs of this peptidase domain are also found in a number of other bacterial genome sequences, most of which are as yet uncharacterized. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFA50087.1 | 5.20e-58 | 1 | 516 | 1 | 506 |
| QNK41697.1 | 2.92e-46 | 343 | 530 | 94 | 283 |
| QBB65500.1 | 8.16e-46 | 350 | 536 | 70 | 258 |
| ALP94484.1 | 9.23e-46 | 350 | 536 | 102 | 290 |
| QEY34222.1 | 5.72e-45 | 343 | 533 | 89 | 277 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NB6_A | 1.36e-37 | 354 | 530 | 22 | 197 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 2OQO_A | 1.88e-37 | 354 | 530 | 22 | 197 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 3DWK_A | 1.29e-30 | 355 | 516 | 30 | 190 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 2OLU_A | 5.06e-30 | 355 | 514 | 39 | 197 | StructuralInsight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme [Staphylococcus aureus],2OLV_A Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus],2OLV_B Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus] |
| 4OON_A | 5.08e-30 | 355 | 532 | 42 | 218 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O66874 | 1.11e-33 | 354 | 537 | 65 | 247 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| P38050 | 1.53e-32 | 339 | 537 | 59 | 251 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| Q04707 | 5.57e-30 | 359 | 530 | 78 | 250 | Penicillin-binding protein 1A OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=ponA PE=1 SV=2 |
| Q00573 | 5.58e-30 | 359 | 530 | 78 | 251 | Penicillin-binding protein 1A (Fragment) OS=Streptococcus oralis OX=1303 GN=ponA PE=3 SV=1 |
| Q8DR59 | 2.40e-29 | 359 | 530 | 78 | 250 | Penicillin-binding protein 1A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=pbpA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000020 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |