You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000056_01839
You are here: Home > Sequence: MGYG000000056_01839
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Muribaculum sp002358615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp002358615 | |||||||||||
| CAZyme ID | MGYG000000056_01839 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61880; End: 64663 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 21 | 680 | 1.3e-62 | 0.699468085106383 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.61e-12 | 105 | 431 | 110 | 434 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.45e-06 | 24 | 147 | 10 | 136 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 2.15e-05 | 75 | 160 | 62 | 151 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10340 | ebgA | 9.21e-05 | 226 | 402 | 245 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD36762.1 | 0.0 | 7 | 927 | 12 | 932 |
| QDH53328.1 | 0.0 | 7 | 927 | 14 | 952 |
| QRM99843.1 | 0.0 | 7 | 908 | 14 | 933 |
| QUT31373.1 | 0.0 | 7 | 908 | 10 | 929 |
| QIU93174.1 | 0.0 | 24 | 927 | 27 | 948 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YPJ_A | 7.32e-10 | 59 | 676 | 53 | 665 | ChainA, Beta galactosidase [Niallia circulans],4YPJ_B Chain B, Beta galactosidase [Niallia circulans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
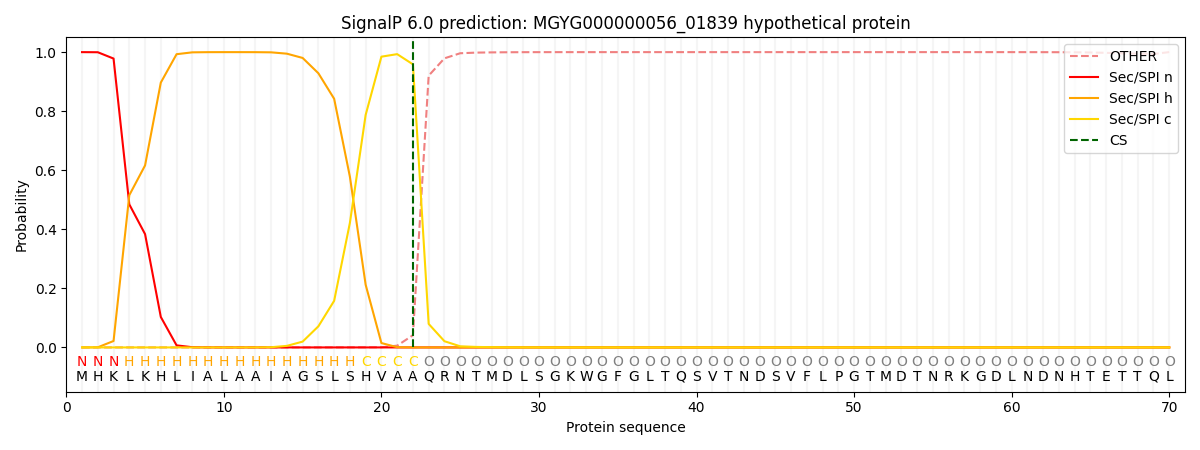
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.008807 | 0.987166 | 0.001380 | 0.000851 | 0.000870 | 0.000896 |
