You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000045_00527
You are here: Home > Sequence: MGYG000000045_00527
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
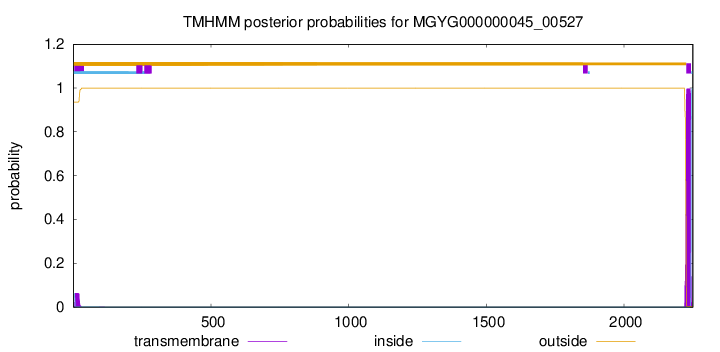
TMHMM annotations
Basic Information help
| Species | Faecalibacillus intestinalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Faecalibacillus; Faecalibacillus intestinalis | |||||||||||
| CAZyme ID | MGYG000000045_00527 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 89499; End: 96251 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 838 | 1277 | 2.3e-108 | 0.9952038369304557 |
| GH3 | 98 | 327 | 1.1e-66 | 0.9768518518518519 |
| CBM6 | 650 | 771 | 7e-20 | 0.8695652173913043 |
| CBM6 | 1447 | 1545 | 1.5e-16 | 0.7246376811594203 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 3.31e-83 | 38 | 452 | 1 | 374 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 1.88e-69 | 42 | 368 | 4 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 3.48e-57 | 94 | 675 | 95 | 693 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 3.95e-36 | 93 | 626 | 67 | 635 | Probable beta-xylosidase; Provisional |
| COG5520 | XynC | 2.39e-34 | 843 | 1278 | 36 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCL58477.1 | 0.0 | 1 | 2249 | 2 | 2250 |
| QQY26973.1 | 0.0 | 1 | 2248 | 1 | 2227 |
| QQV06088.1 | 0.0 | 1 | 2248 | 1 | 2227 |
| QPS14204.1 | 0.0 | 1 | 2248 | 1 | 2227 |
| QMW75461.1 | 0.0 | 1 | 2248 | 1 | 2227 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M6G_A | 6.96e-135 | 22 | 622 | 42 | 612 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 6JGC_A | 9.48e-117 | 22 | 622 | 11 | 601 | Crystalstructure of barley exohydrolaseI W286Y mutant in complex with glucose. [Hordeum vulgare subsp. vulgare],6JGD_A Crystal structure of barley exohydrolaseI W286Y mutant in complex with methyl 6-thio-beta-gentiobioside [Hordeum vulgare subsp. vulgare] |
| 6JG7_A | 3.23e-116 | 22 | 622 | 11 | 601 | Crystalstructure of barley exohydrolaseI W286F in complex with methyl 2-thio-beta-sophoroside [Hordeum vulgare subsp. vulgare],6JGA_A Crystal structure of barley exohydrolaseI W286F in complex with 4'-nitrophenyl thiolaminaribioside [Hordeum vulgare subsp. vulgare],6JGB_A Crystal structure of barley exohydrolaseI W286F mutant in complex with methyl 6-thio-beta-gentiobioside [Hordeum vulgare subsp. vulgare] |
| 1LQ2_A | 3.58e-116 | 22 | 622 | 7 | 597 | Crystalstructure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X38_A crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X39_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare] |
| 1EX1_A | 3.91e-116 | 22 | 622 | 7 | 597 | Beta-d-glucanExohydrolase From Barley [Hordeum vulgare],1IEQ_A Crystal Structure Of Barley Beta-D-Glucan Glucohydrolase Isoenzyme Exo1 [Hordeum vulgare],1IEV_A Crystal Structure Of Barley Beta-d-glucan Glucohydrolase Isoenzyme Exo1 In Complex With Cyclohexitol [Hordeum vulgare],1IEW_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 2-deoxy-2-fluoro-alpha-D-glucoside [Hordeum vulgare],1IEX_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4I,4III,4V-S-trithiocellohexaose [Hordeum vulgare],1J8V_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4'-nitrophenyl 3I-thiolaminaritrioside [Hordeum vulgare],3WLH_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],3WLJ_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 3-deoxy-glucose [Hordeum vulgare subsp. vulgare],3WLK_X Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 4-deoxy-glucose [Hordeum vulgare subsp. vulgare],3WLL_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with PEG400 [Hordeum vulgare subsp. vulgare],3WLM_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with octyl-O-glucoside [Hordeum vulgare subsp. vulgare],3WLN_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with octyl-S-glucoside [Hordeum vulgare subsp. vulgare],3WLP_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q23892 | 7.21e-58 | 31 | 608 | 80 | 677 | Lysosomal beta glucosidase OS=Dictyostelium discoideum OX=44689 GN=gluA PE=1 SV=2 |
| A7LXU3 | 3.71e-55 | 60 | 622 | 85 | 657 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
| T2KMH0 | 2.84e-45 | 101 | 733 | 64 | 703 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
| Q56078 | 1.87e-44 | 102 | 675 | 104 | 693 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
| P33363 | 2.49e-44 | 102 | 675 | 104 | 693 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.999072 | 0.000179 | 0.000159 | 0.000157 | 0.000143 |