You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004869_03009
You are here: Home > Sequence: MGYG000004869_03009
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
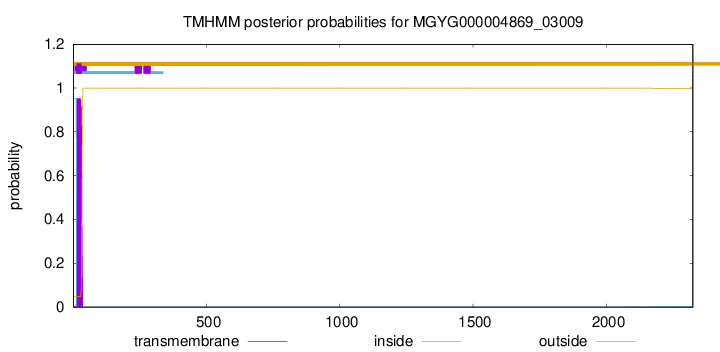
TMHMM annotations
Basic Information help
| Species | Lactonifactor sp009677585 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lactonifactor; Lactonifactor sp009677585 | |||||||||||
| CAZyme ID | MGYG000004869_03009 | |||||||||||
| CAZy Family | CBM66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 229154; End: 236131 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH43 | 1575 | 1864 | 2.2e-102 | 0.9965986394557823 |
| GH43 | 318 | 581 | 3.5e-92 | 0.9888059701492538 |
| GH43 | 855 | 1080 | 3.4e-55 | 0.9856459330143541 |
| CBM66 | 135 | 295 | 1.1e-27 | 0.9354838709677419 |
| CBM35 | 1361 | 1482 | 2.6e-16 | 0.9747899159663865 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd18820 | GH43_LbAraf43-like | 2.70e-122 | 1584 | 1839 | 1 | 258 | Glycosyl hydrolase family 43 proteins similar to Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans GbtXyl43B. This uncharacterized glycosyl hydrolase family 43 (GH43) subgroup belongs to a subgroup which includes enzymes with beta-xylosidase (EC 3.2.1.37), alpha-L-arabinofuranosidase (EC 3.2.1.55) and possibly bifunctional xylosidase/arabinofuranosidase activities, similar to Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans IT-08 beta-xylosidase / exo-xylanase (GbtXyl43B). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08991 | GH43_HoAraf43-like | 2.01e-117 | 319 | 597 | 1 | 283 | Glycosyl hydrolase family 43 protein such as Halothermothrix orenii H 168 alpha-L-arabinofuranosidase (HoAraf43;Hore_20580). This glycosyl hydrolase family 43 (GH43) subgroup includes Halothermothrix orenii H 168 alpha-L-arabinofuranosidase (EC 3.2.1.55) (HoAraf43;Hore_20580). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. This GH43_ HoAraf43-like subgroup includes enzymes that have been annotated as having xylan-digesting beta-xylosidase (EC 3.2.1.37) and xylanase (endo-alpha-L-arabinanase, EC 3.2.1.8) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| NF033930 | pneumo_PspA | 2.65e-93 | 2106 | 2321 | 443 | 659 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| NF033930 | pneumo_PspA | 6.60e-87 | 2065 | 2265 | 442 | 643 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| NF033930 | pneumo_PspA | 2.10e-82 | 2045 | 2254 | 442 | 652 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNU68671.1 | 4.74e-195 | 662 | 1876 | 305 | 1366 |
| AUG57492.1 | 3.08e-186 | 707 | 1876 | 37 | 1059 |
| BBA94052.1 | 2.10e-183 | 658 | 1876 | 301 | 1366 |
| AEY66127.1 | 7.38e-183 | 702 | 1876 | 4 | 1029 |
| QGH33897.1 | 6.59e-160 | 694 | 1350 | 21 | 651 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3KST_A | 4.54e-63 | 319 | 601 | 25 | 303 | Crystalstructure of Endo-1,4-beta-xylanase (NP_811807.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.70 A resolution [Bacteroides thetaiotaomicron VPI-5482],3KST_B Crystal structure of Endo-1,4-beta-xylanase (NP_811807.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.70 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 4QQS_A | 1.14e-56 | 308 | 604 | 3 | 312 | Crystalstructure of a thermostable family-43 glycoside hydrolase [Halothermothrix orenii H 168],4QQS_B Crystal structure of a thermostable family-43 glycoside hydrolase [Halothermothrix orenii H 168] |
| 5M8B_A | 9.18e-50 | 1575 | 1875 | 27 | 346 | ChainA, Alpha-L-arabinofuranosidase II [Levilactobacillus brevis],5M8B_B Chain B, Alpha-L-arabinofuranosidase II [Levilactobacillus brevis] |
| 3K1U_A | 1.26e-46 | 1577 | 1879 | 11 | 327 | Beta-xylosidase,family 43 glycosyl hydrolase from Clostridium acetobutylicum [Clostridium acetobutylicum] |
| 3AKF_A | 3.03e-46 | 1572 | 1871 | 8 | 317 | Crystalstructure of exo-1,5-alpha-L-arabinofuranosidase [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKG_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranobiose [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKH_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranotriose [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKI_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido [Streptomyces avermitilis MA-4680 = NBRC 14893] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82P90 | 2.18e-45 | 1572 | 1871 | 34 | 343 | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=Araf43A PE=1 SV=1 |
| P82594 | 1.49e-37 | 1575 | 1838 | 50 | 318 | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| G4MMH2 | 4.24e-35 | 1574 | 1870 | 40 | 348 | Alpha-L-arabinofuranosidase B OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=abfB PE=1 SV=1 |
| A0A3R0A696 | 9.87e-33 | 727 | 1095 | 377 | 758 | Alpha-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=blArafA PE=1 SV=1 |
| P48791 | 3.33e-13 | 319 | 588 | 13 | 314 | Beta-xylosidase OS=Prevotella ruminicola OX=839 GN=xynB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
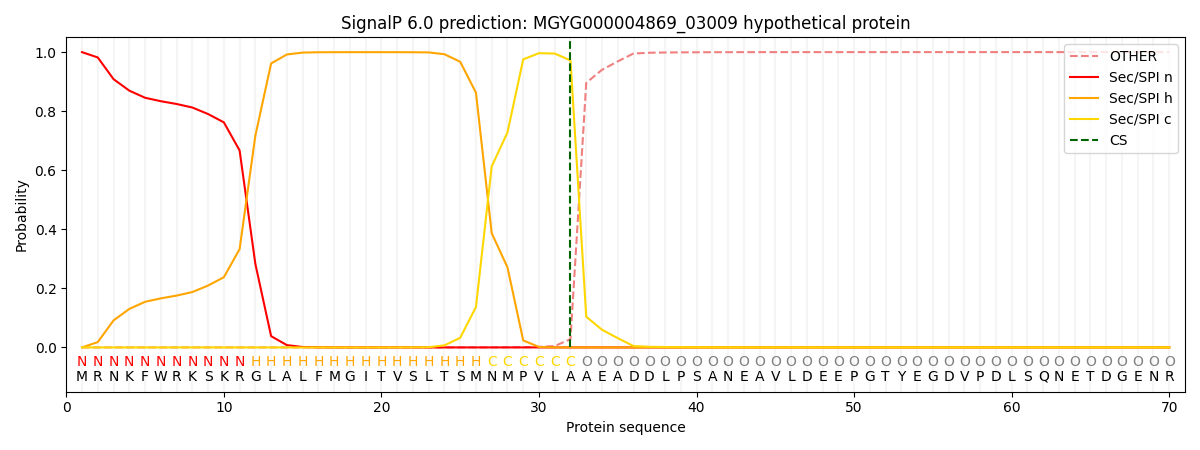
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000491 | 0.998522 | 0.000342 | 0.000242 | 0.000195 | 0.000163 |