You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004726_01759
You are here: Home > Sequence: MGYG000004726_01759
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
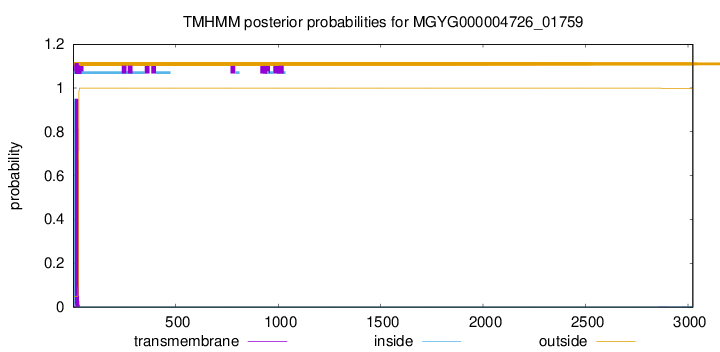
TMHMM annotations
Basic Information help
| Species | Pseudoflavonifractor sp014287675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Pseudoflavonifractor; Pseudoflavonifractor sp014287675 | |||||||||||
| CAZyme ID | MGYG000004726_01759 | |||||||||||
| CAZy Family | CBM66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 349336; End: 358410 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 1707 | 1906 | 1.1e-25 | 0.8854625550660793 |
| CBM66 | 409 | 549 | 5.4e-18 | 0.8709677419354839 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 1.78e-34 | 1661 | 1924 | 131 | 385 | Predicted peptidase [General function prediction only]. |
| pfam03629 | SASA | 9.36e-23 | 2222 | 2456 | 6 | 223 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
| cd02850 | E_set_Cellulase_N | 4.11e-13 | 720 | 795 | 1 | 86 | N-terminal Early set domain associated with the catalytic domain of cellulase. E or "early" set domains are associated with the catalytic domain of cellulases at the N-terminal end. Cellulases are O-glycosyl hydrolases (GHs) that hydrolyze beta 1-4 glucosidic bonds in cellulose. They are usually categorized into either exoglucanases, which sequentially release terminal sugar units from the cellulose chain, or endoglucanases, which also attack the chain internally. The N-terminal domain of cellulase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| COG5492 | YjdB | 1.58e-12 | 1347 | 1504 | 179 | 319 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 4.73e-12 | 1352 | 1427 | 3 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG58036.1 | 8.97e-187 | 450 | 1343 | 104 | 961 |
| SDT49439.1 | 1.29e-99 | 709 | 1350 | 221 | 846 |
| BCI61582.1 | 1.81e-43 | 1690 | 1925 | 809 | 1042 |
| QDU56037.1 | 1.44e-31 | 1691 | 1928 | 785 | 1009 |
| QTH41961.1 | 2.61e-25 | 2694 | 3023 | 1990 | 2338 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 3.93e-32 | 1681 | 1926 | 133 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 3.01e-28 | 1709 | 1932 | 23 | 222 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 2.01e-13 | 2851 | 3020 | 1681 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 2.20e-13 | 2847 | 3020 | 903 | 1082 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| C6CRV0 | 2.07e-10 | 2680 | 3017 | 1074 | 1457 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P49052 | 2.95e-08 | 2827 | 3012 | 9 | 197 | S-layer protein OS=Bacillus licheniformis OX=1402 PE=3 SV=1 |
| P94217 | 2.55e-07 | 2827 | 3012 | 9 | 197 | S-layer protein EA1 OS=Bacillus anthracis OX=1392 GN=eag PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
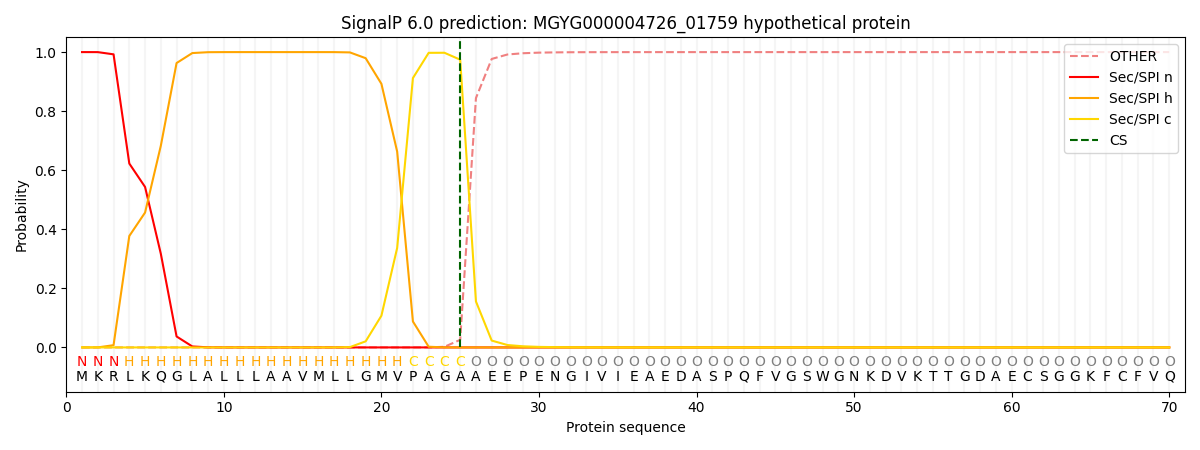
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000275 | 0.999087 | 0.000166 | 0.000172 | 0.000157 | 0.000147 |