You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004703_00471
You are here: Home > Sequence: MGYG000004703_00471
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900556275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900556275 | |||||||||||
| CAZyme ID | MGYG000004703_00471 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 23961; End: 25820 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 145 | 514 | 4.7e-45 | 0.9151670951156813 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03901 | Glyco_transf_22 | 2.60e-19 | 147 | 499 | 4 | 360 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| PLN02816 | PLN02816 | 5.78e-13 | 162 | 515 | 59 | 425 | mannosyltransferase |
| pfam04138 | GtrA | 7.84e-12 | 18 | 125 | 1 | 114 | GtrA-like protein. Members of this family are predicted to be integral membrane proteins with three or four transmembrane spans. They are involved in the synthesis of cell surface polysaccharides. The GtrA family are a subset of this family. GtrA is predicted to be an integral membrane protein with 4 transmembrane spans. It is involved is in O antigen modification by Shigella flexneri bacteriophage X (SfX), but does not determine the specificity of glucosylation. Its function remains unknown, but it may play a role in translocation of undecaprenyl phosphate linked glucose (UndP-Glc) across the cytoplasmic membrane. Another member of this family is a DTDP-glucose-4-keto-6-deoxy-D-glucose reductase, which catalyzes the conversion of dTDP-4-keto-6-deoxy-D-glucose to dTDP-D-fucose, which is involved in the biosynthesis of the serotype-specific polysaccharide antigen of Actinobacillus actinomycetemcomitans Y4 (serotype b). This family also includes the teichoic acid glycosylation protein, GtcA, which is a serotype-specific protein in some Listeria innocua and monocytogenes strains. Its exact function is not known, but it is essential for decoration of cell wall teichoic acids with glucose and galactose. |
| COG2246 | GtrA | 4.30e-07 | 15 | 124 | 12 | 131 | Putative flippase GtrA (transmembrane translocase of bactoprenol-linked glucose) [Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEW21902.1 | 9.00e-72 | 143 | 616 | 27 | 499 |
| QDW24341.1 | 2.09e-68 | 151 | 616 | 22 | 490 |
| AOW16884.1 | 1.38e-66 | 149 | 600 | 16 | 474 |
| QVY67119.1 | 2.12e-66 | 147 | 619 | 14 | 490 |
| QHS61266.1 | 1.07e-63 | 146 | 617 | 10 | 474 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6CAB8 | 4.21e-11 | 145 | 522 | 19 | 382 | GPI mannosyltransferase 3 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=GPI10 PE=3 SV=1 |
| Q1LZA0 | 4.18e-10 | 126 | 454 | 36 | 355 | GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1 |
| Q94A15 | 9.74e-10 | 162 | 465 | 59 | 353 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
| Q92521 | 1.71e-09 | 144 | 474 | 63 | 382 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
| Q9JJQ0 | 2.94e-09 | 149 | 513 | 57 | 407 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000021 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
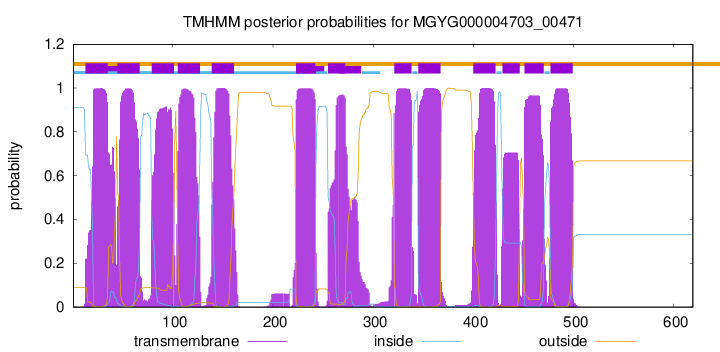
| start | end |
|---|---|
| 13 | 35 |
| 45 | 67 |
| 79 | 101 |
| 105 | 127 |
| 139 | 161 |
| 223 | 242 |
| 255 | 272 |
| 321 | 338 |
| 345 | 367 |
| 400 | 422 |
| 429 | 446 |
| 451 | 470 |
| 477 | 499 |
