You are browsing environment: HUMAN GUT
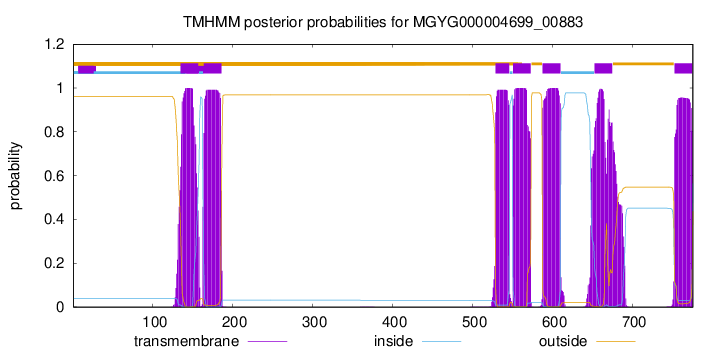
MGYG000004699_00883
Basic Information
help
Species
Lineage
Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium;
CAZyme ID
MGYG000004699_00883
CAZy Family
GT84
CAZyme Description
Cyclic beta-(1,2)-glucan synthase NdvB
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
776
90037.62
7.6034
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004699
3820928
MAG
Germany
Europe
Gene Location
Start: 1;
End: 2331
Strand: +
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam13632
Glyco_trans_2_3
7.33e-04
346
541
1
180
Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
P20471
1.84e-110
12
769
282
1059
Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2
more
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000060
0.000000
0.000000
0.000000
0.000000
0.000000
start
end
135
157
164
186
529
546
551
573
588
610
653
675
753
775