You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004676_01093
You are here: Home > Sequence: MGYG000004676_01093
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900766005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900766005 | |||||||||||
| CAZyme ID | MGYG000004676_01093 | |||||||||||
| CAZy Family | PL13 | |||||||||||
| CAZyme Description | Heparin lyase I | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 58142; End: 59317 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL13 | 22 | 391 | 2.7e-196 | 0.9944903581267218 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14099 | Polysacc_lyase | 1.10e-14 | 60 | 378 | 18 | 200 | Polysaccharide lyase. This family includes heparin lyase I, EC:4.2.2.7. Heparin lyase I depolymerizes heparin by cleaving the glycosidic linkage next to an iduronic acid moiety. The structure of heparin lyase I consists of a beta-jelly roll domain with a long, deep substrate-binding groove and an unusual thumb domain containing many basic residues extending from the main body of the enzyme. This family also includes glucuronan lyase, EC:4.2.2.14. The structure glucuronan lyase is a beta-jelly roll. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU95134.1 | 1.33e-244 | 1 | 391 | 1 | 392 |
| QRQ56254.1 | 3.31e-238 | 1 | 391 | 1 | 392 |
| SCV08896.1 | 3.31e-238 | 1 | 391 | 1 | 392 |
| ALJ49472.1 | 3.31e-238 | 1 | 391 | 1 | 392 |
| QUT82562.1 | 4.71e-238 | 1 | 391 | 1 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3IKW_A | 3.64e-232 | 18 | 391 | 1 | 374 | Structureof Heparinase I from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron] |
| 3IMN_A | 9.90e-231 | 16 | 391 | 1 | 376 | Crystalstructure of heparin lyase I from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],3IN9_A Crystal structure of heparin lyase I complexed with disaccharide heparin [Bacteroides thetaiotaomicron] |
| 3ILR_A | 2.11e-230 | 27 | 391 | 6 | 370 | Structureof Heparinase I from Bacteroides thetaiotaomicron in complex with tetrasaccharide product [Bacteroides thetaiotaomicron] |
| 3INA_A | 3.30e-229 | 16 | 391 | 1 | 376 | Crystalstructure of heparin lyase I H151A mutant complexed with a dodecasaccharide heparin [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q05819 | 8.35e-170 | 1 | 390 | 1 | 381 | Heparin lyase I OS=Pedobacter heparinus OX=984 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
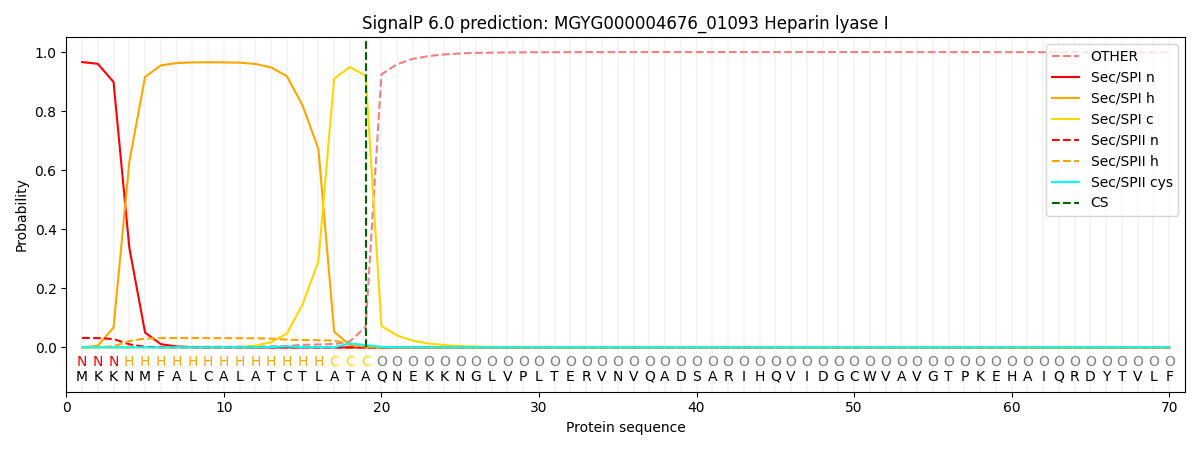
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004069 | 0.960910 | 0.033980 | 0.000475 | 0.000279 | 0.000268 |
