You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004658_02007
You are here: Home > Sequence: MGYG000004658_02007
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes finegoldii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes finegoldii | |||||||||||
| CAZyme ID | MGYG000004658_02007 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48108; End: 50582 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 56 | 264 | 1.2e-72 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 2.21e-101 | 20 | 810 | 27 | 751 | beta-glucosidase BglX. |
| COG1472 | BglX | 1.64e-74 | 53 | 372 | 51 | 376 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PLN03080 | PLN03080 | 5.21e-55 | 21 | 783 | 48 | 742 | Probable beta-xylosidase; Provisional |
| pfam00933 | Glyco_hydro_3 | 2.94e-46 | 59 | 296 | 63 | 314 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 6.63e-38 | 519 | 690 | 53 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFL78920.1 | 0.0 | 1 | 824 | 1 | 824 |
| BBL13639.1 | 0.0 | 1 | 822 | 1 | 820 |
| BBL04227.1 | 0.0 | 1 | 822 | 1 | 820 |
| BBL13665.1 | 0.0 | 20 | 818 | 3 | 796 |
| BBL05223.1 | 0.0 | 16 | 818 | 17 | 814 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7MS2_A | 6.20e-122 | 32 | 822 | 7 | 666 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 3AC0_A | 1.74e-119 | 32 | 816 | 8 | 828 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 3ABZ_A | 1.50e-115 | 32 | 816 | 8 | 828 | Crystalstructure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_B Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_C Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_D Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus] |
| 4I3G_A | 1.41e-107 | 4 | 824 | 28 | 826 | CrystalStructure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae],4I3G_B Crystal Structure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae] |
| 2X40_A | 7.55e-101 | 28 | 820 | 2 | 709 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NDE2 | 6.42e-134 | 34 | 816 | 10 | 823 | Probable beta-glucosidase I OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglI PE=3 SV=2 |
| Q2U8Y5 | 6.42e-134 | 34 | 816 | 10 | 823 | Probable beta-glucosidase I OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglI PE=3 SV=1 |
| A1CA51 | 7.19e-132 | 36 | 816 | 12 | 822 | Probable beta-glucosidase I OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=bglI PE=3 SV=1 |
| A1DFA8 | 1.01e-131 | 34 | 816 | 10 | 822 | Probable beta-glucosidase I OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=bglI PE=3 SV=1 |
| Q0CAF5 | 2.04e-131 | 34 | 816 | 10 | 823 | Probable beta-glucosidase I OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=bglI PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
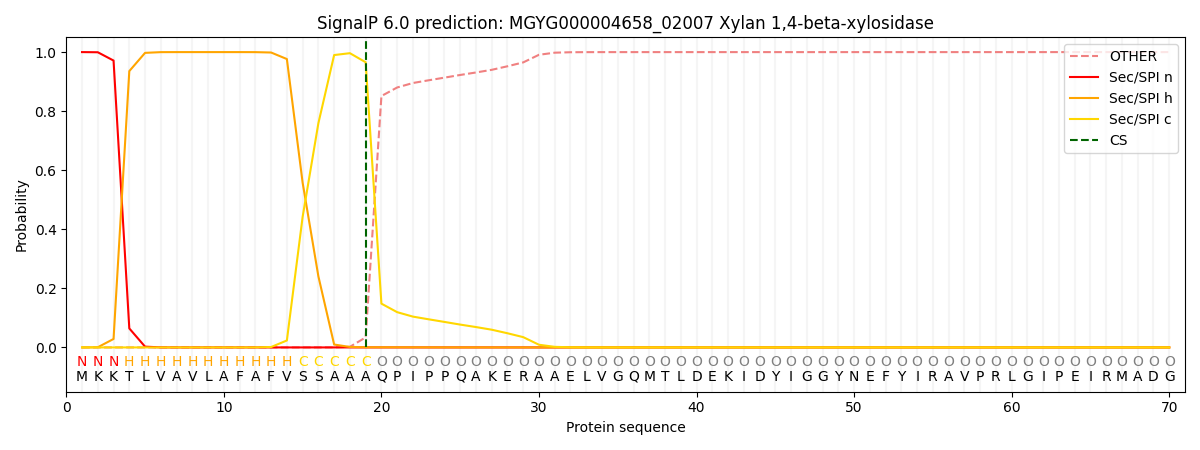
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000241 | 0.999143 | 0.000155 | 0.000162 | 0.000150 | 0.000135 |
