You are browsing environment: HUMAN GUT
MGYG000004621_00324
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella;
CAZyme ID
MGYG000004621_00324
CAZy Family
PL35
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004621
2838817
MAG
France
Europe
Gene Location
Start: 31618;
End: 33498
Strand: -
No EC number prediction in MGYG000004621_00324.
Family
Start
End
Evalue
family coverage
PL35
410
578
3.1e-50
0.9608938547486033
MGYG000004621_00324 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
3A0O_A
8.16e-09
37
274
121
373
Crystalstructure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58],3A0O_B Crystal structure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58]
more
3AFL_A
8.16e-09
37
274
121
373
Crystalstructure of exotype alginate lyase Atu3025 H531A complexed with alginate trisaccharide [Agrobacterium fabrum str. C58]
more
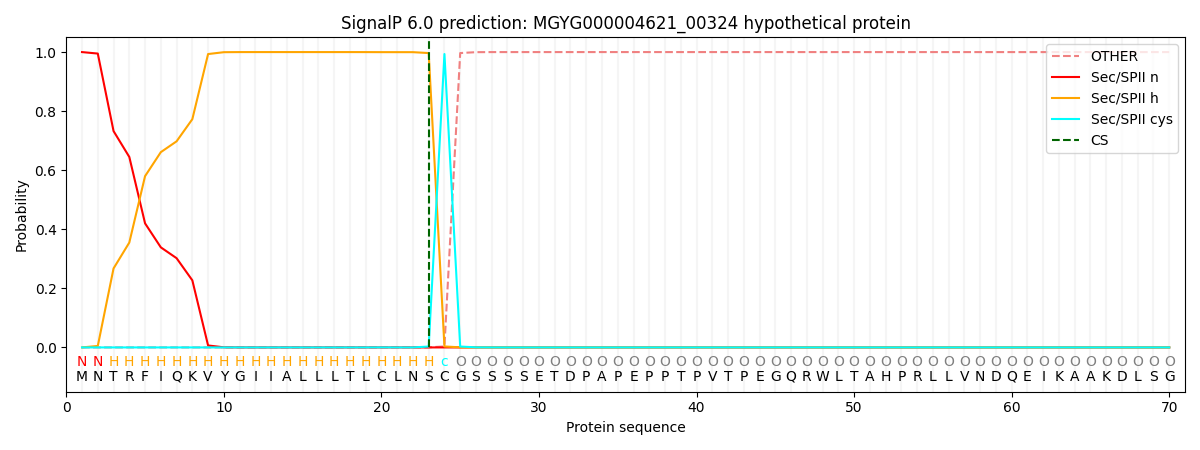
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000002
1.000049
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000004621_00324.