You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004490_00988
Basic Information
help
| Species |
Muribaculum sp003150235
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp003150235
|
| CAZyme ID |
MGYG000004490_00988
|
| CAZy Family |
GT83 |
| CAZyme Description |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 581 |
|
65978.79 |
8.8009 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004490 |
3323418 |
MAG |
Israel |
Asia |
|
| Gene Location |
Start: 18530;
End: 20275
Strand: +
|
No EC number prediction in MGYG000004490_00988.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
5 |
470 |
2.7e-77 |
0.812962962962963 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.61e-25 |
8 |
466 |
7 |
433 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| COG4745
|
COG4745 |
0.001 |
13 |
206 |
20 |
202 |
Predicted membrane-bound mannosyltransferase [General function prediction only]. |
| pfam13231
|
PMT_2 |
0.004 |
65 |
226 |
2 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O67270
|
1.83e-14 |
14 |
493 |
6 |
423 |
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
|
Q7N3Q9
|
1.60e-06 |
1 |
459 |
1 |
423 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Photorhabdus laumondii subsp. laumondii (strain DSM 15139 / CIP 105565 / TT01) OX=243265 GN=arnT PE=2 SV=1 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999670
|
0.000275
|
0.000042
|
0.000002
|
0.000001
|
0.000037
|
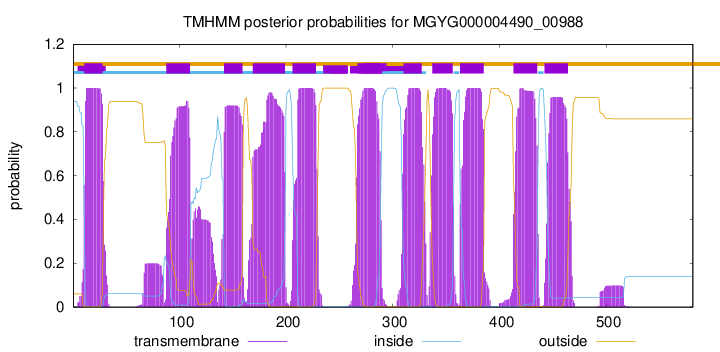
| start |
end |
| 11 |
28 |
| 88 |
110 |
| 142 |
159 |
| 169 |
199 |
| 206 |
228 |
| 267 |
289 |
| 310 |
327 |
| 337 |
356 |
| 363 |
385 |
| 413 |
435 |
| 442 |
464 |


