You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004468_00612
You are here: Home > Sequence: MGYG000004468_00612
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A sp900539755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A sp900539755 | |||||||||||
| CAZyme ID | MGYG000004468_00612 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 748868; End: 751744 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 47 | 958 | 2.9e-194 | 0.9939320388349514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 2.87e-43 | 210 | 770 | 351 | 866 | alpha-L-rhamnosidase. |
| NF038009 | TatB_1 | 0.002 | 1 | 52 | 14 | 70 | twin-arginine translocation system-associated protein. This family is suggested by TCDB to be part of the twin-arginine translocation (TAT) system, but lacks detectable sequence similarity to subunits such as TatB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG53460.1 | 0.0 | 12 | 958 | 1 | 947 |
| BCG53464.1 | 0.0 | 12 | 958 | 1 | 954 |
| BBD44720.1 | 0.0 | 1 | 958 | 1 | 954 |
| SCM56507.1 | 0.0 | 1 | 958 | 1 | 953 |
| BAR48036.1 | 0.0 | 1 | 958 | 2 | 956 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 1.23e-28 | 252 | 957 | 457 | 1138 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 2.19e-26 | 216 | 955 | 378 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 4.99e-26 | 216 | 955 | 378 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 4.54e-35 | 62 | 799 | 46 | 743 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
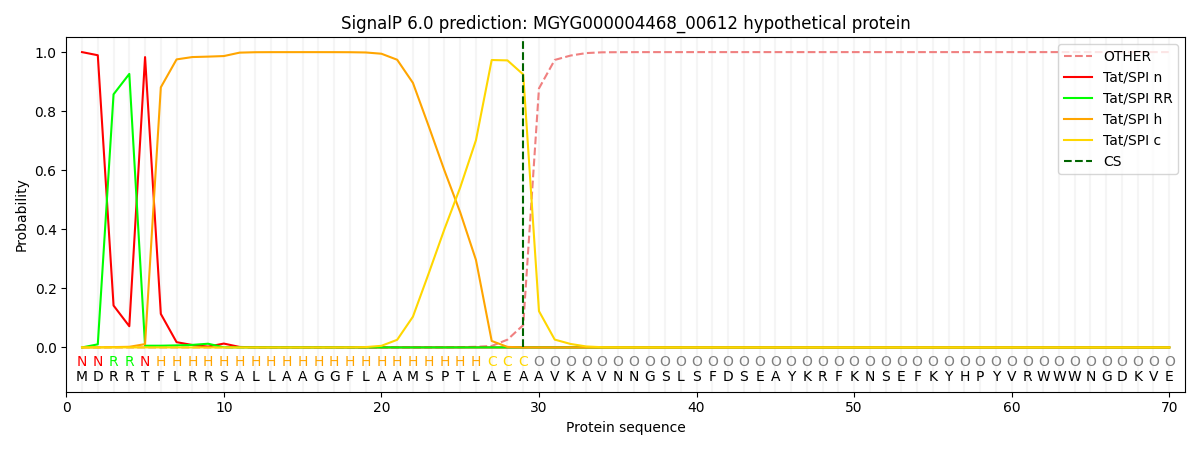
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999985 | 0.000010 | 0.000000 |
