You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004431_00706
You are here: Home > Sequence: MGYG000004431_00706
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | COE1 sp001916965 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; COE1; COE1 sp001916965 | |||||||||||
| CAZyme ID | MGYG000004431_00706 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase Y | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26551; End: 27831 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 86 | 417 | 1.5e-81 | 0.9834983498349835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 2.82e-84 | 89 | 415 | 5 | 307 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 5.05e-75 | 128 | 415 | 2 | 262 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 8.58e-60 | 92 | 414 | 32 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADX05700.1 | 5.89e-106 | 81 | 426 | 63 | 409 |
| ADU22101.1 | 1.98e-87 | 84 | 417 | 53 | 380 |
| AEE64767.1 | 9.00e-82 | 85 | 417 | 41 | 367 |
| BAF93203.1 | 1.38e-79 | 47 | 418 | 23 | 391 |
| AHF24514.1 | 1.04e-75 | 56 | 414 | 11 | 364 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NL2_A | 1.68e-57 | 84 | 426 | 11 | 347 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 2W5F_A | 6.45e-54 | 72 | 413 | 176 | 521 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 3W24_A | 7.40e-54 | 93 | 426 | 17 | 334 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W27_A | 5.64e-53 | 93 | 426 | 17 | 334 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W28_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W29_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W25_A | 5.64e-53 | 93 | 426 | 17 | 334 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W26_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26223 | 1.67e-60 | 83 | 422 | 1 | 344 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| P29126 | 7.19e-55 | 84 | 413 | 629 | 946 | Bifunctional endo-1,4-beta-xylanase XylA OS=Ruminococcus flavefaciens OX=1265 GN=xynA PE=3 SV=1 |
| P51584 | 4.82e-52 | 72 | 413 | 187 | 532 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q60042 | 1.35e-50 | 71 | 425 | 352 | 694 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| Q60037 | 1.56e-49 | 71 | 425 | 356 | 698 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
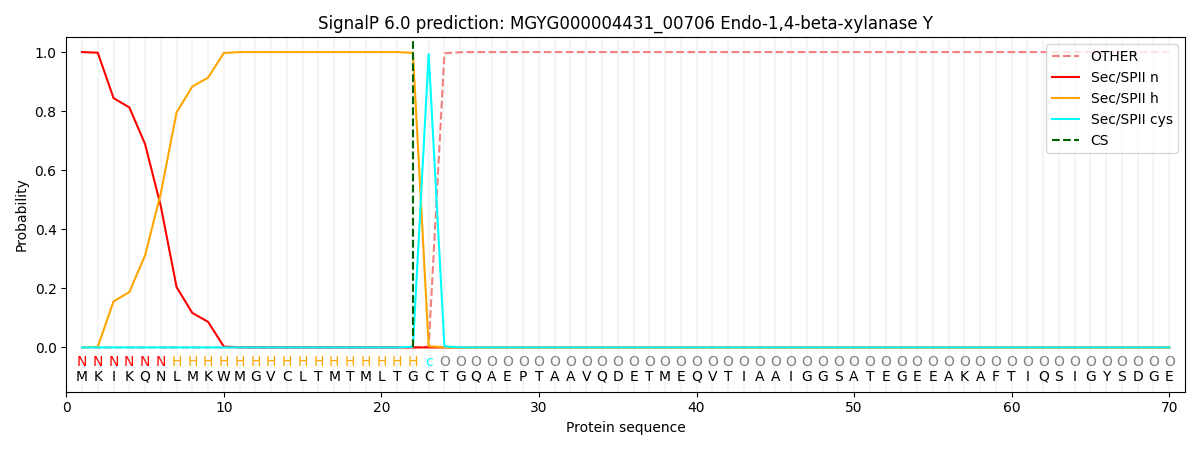
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000068 | 0.000000 | 0.000000 | 0.000000 |
