You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004393_01407
You are here: Home > Sequence: MGYG000004393_01407
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
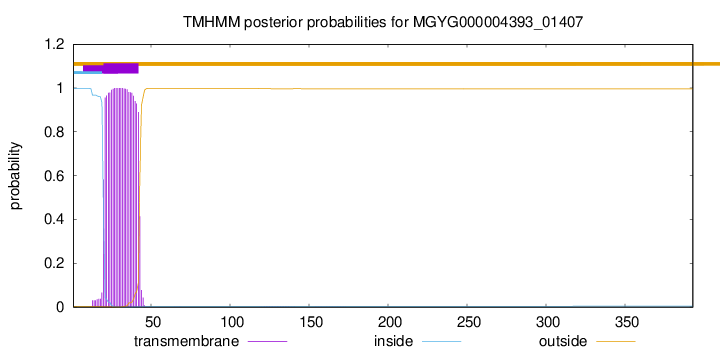
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-460; | |||||||||||
| CAZyme ID | MGYG000004393_01407 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6004; End: 7185 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 3.42e-30 | 276 | 388 | 3 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG0791 | Spr | 5.48e-23 | 253 | 384 | 67 | 195 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| NF033742 | NlpC_p60_RipB | 5.17e-18 | 261 | 392 | 77 | 206 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
| PRK10838 | spr | 5.31e-18 | 294 | 393 | 93 | 188 | bifunctional murein DD-endopeptidase/murein LD-carboxypeptidase. |
| NF033741 | NlpC_p60_RipA | 3.05e-14 | 276 | 360 | 342 | 428 | NlpC/P60 family peptidoglycan endopeptidase RipA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFS33417.1 | 8.40e-185 | 8 | 391 | 2 | 387 |
| QIF80185.1 | 8.40e-185 | 8 | 391 | 2 | 387 |
| CEK36621.1 | 4.08e-174 | 6 | 391 | 9 | 392 |
| CEJ75496.1 | 1.17e-173 | 6 | 391 | 9 | 392 |
| CEK40080.1 | 1.65e-173 | 6 | 391 | 9 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2K1G_A | 1.44e-13 | 277 | 390 | 21 | 124 | SolutionNMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 [Escherichia coli K-12] |
| 7CFL_A | 3.49e-12 | 262 | 390 | 15 | 138 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 3NE0_A | 1.69e-10 | 262 | 360 | 86 | 185 | Structureand functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation [Mycobacterium tuberculosis H37Rv] |
| 3PBC_A | 1.69e-10 | 262 | 360 | 86 | 185 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
| 4Q4T_A | 7.99e-10 | 262 | 360 | 344 | 443 | Structureof the Resuscitation Promoting Factor Interacting protein RipA mutated at E444 [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q65NQ9 | 4.52e-48 | 247 | 391 | 310 | 452 | Peptidoglycan DL-endopeptidase CwlO OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=cwlO PE=3 SV=1 |
| P40767 | 5.09e-46 | 261 | 391 | 343 | 473 | Peptidoglycan DL-endopeptidase CwlO OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlO PE=1 SV=2 |
| P45296 | 2.54e-13 | 226 | 390 | 42 | 183 | Probable endopeptidase NlpC homolog OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=nlpC PE=3 SV=1 |
| P0AFV7 | 9.68e-13 | 235 | 390 | 31 | 185 | Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase OS=Shigella flexneri OX=623 GN=mepS PE=3 SV=1 |
| P0AFV5 | 9.68e-13 | 235 | 390 | 31 | 185 | Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=mepS PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
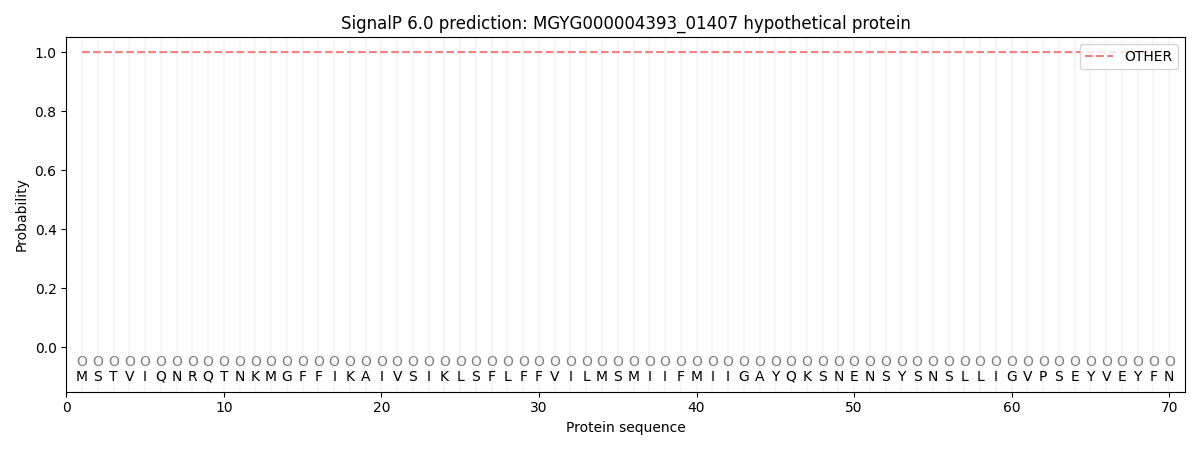
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999997 | 0.000032 | 0.000000 | 0.000000 | 0.000000 | 0.000003 |