You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004281_00854
You are here: Home > Sequence: MGYG000004281_00854
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
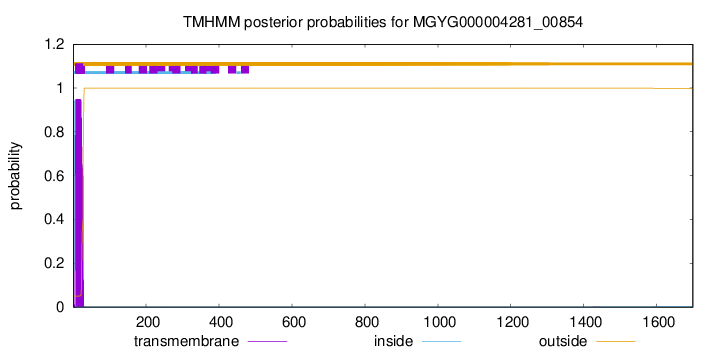
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900552465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900552465 | |||||||||||
| CAZyme ID | MGYG000004281_00854 | |||||||||||
| CAZy Family | GH20 | |||||||||||
| CAZyme Description | N-acetylneuraminate epimerase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67494; End: 72593 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 142 | 412 | 5.7e-61 | 0.9643916913946587 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03548 | mutarot_permut | 3.60e-113 | 1365 | 1691 | 3 | 331 | cyclically-permuted mutarotase family protein. Members of this protein family show essentially full-length homology, cyclically permuted, to YjhT from Escherichia coli. YjhT was shown to act as a mutarotase for sialic acid, and by this ability to be able to act as a virulence factor. Members of the YjhT family (TIGR03547) and this cyclically-permuted family have multiple repeats of the beta-propeller-forming Kelch repeat. |
| cd02742 | GH20_hexosaminidase | 3.49e-93 | 147 | 412 | 1 | 303 | Beta-N-acetylhexosaminidases of glycosyl hydrolase family 20 (GH20) catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. These enzymes are broadly distributed in microorganisms, plants and animals, and play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, viral penetration, the development of carcinomas, inflammatory events and lysosomal storage disorders. The GH20 enzymes include the eukaryotic beta-N-acetylhexosaminidases A and B, the bacterial chitobiases, dispersin B, and lacto-N-biosidase. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself. |
| cd01827 | sialate_O-acetylesterase_like1 | 3.84e-76 | 677 | 868 | 1 | 188 | sialate O-acetylesterase_like family of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| cd06563 | GH20_chitobiase-like | 4.34e-50 | 145 | 444 | 1 | 357 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| pfam00728 | Glyco_hydro_20 | 2.34e-45 | 145 | 411 | 1 | 343 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50180.1 | 0.0 | 1 | 667 | 1 | 667 |
| QIX64921.1 | 0.0 | 1 | 667 | 1 | 669 |
| QUT52022.1 | 0.0 | 1 | 667 | 1 | 669 |
| QUR48261.1 | 0.0 | 1 | 667 | 1 | 669 |
| QUT21124.1 | 0.0 | 24 | 667 | 24 | 669 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KMM_A | 2.81e-55 | 876 | 1343 | 25 | 634 | ChainA, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306],7KMM_B Chain B, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306] |
| 4PYS_A | 1.22e-36 | 93 | 467 | 78 | 505 | Thecrystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343 [Bacteroides fragilis NCTC 9343],4PYS_B The crystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343 [Bacteroides fragilis NCTC 9343] |
| 6YHH_A | 1.60e-36 | 25 | 411 | 8 | 476 | X-rayStructure of Flavobacterium johnsoniae chitobiase (FjGH20) [Flavobacterium johnsoniae UW101],6YHH_B X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) [Flavobacterium johnsoniae UW101] |
| 7DUP_A | 8.06e-36 | 67 | 284 | 59 | 316 | ChainA, Beta-N-acetylhexosaminidase [Bacteroides thetaiotaomicron],7DVA_A Chain A, Beta-N-acetylhexosaminidase [Bacteroides thetaiotaomicron],7DVA_B Chain B, Beta-N-acetylhexosaminidase [Bacteroides thetaiotaomicron] |
| 7DVB_A | 3.50e-35 | 67 | 284 | 59 | 316 | ChainA, Beta-N-acetylhexosaminidase [Bacteroides thetaiotaomicron],7DVB_B Chain B, Beta-N-acetylhexosaminidase [Bacteroides thetaiotaomicron],7DVB_C Chain C, Beta-N-acetylhexosaminidase [Bacteroides thetaiotaomicron],7DVB_D Chain D, Beta-N-acetylhexosaminidase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82450 | 6.01e-37 | 879 | 1321 | 30 | 509 | Sialate O-acetylesterase OS=Rattus norvegicus OX=10116 GN=Siae PE=1 SV=2 |
| P70665 | 5.07e-34 | 879 | 1285 | 30 | 468 | Sialate O-acetylesterase OS=Mus musculus OX=10090 GN=Siae PE=1 SV=3 |
| Q9HAT2 | 5.32e-34 | 873 | 1304 | 25 | 464 | Sialate O-acetylesterase OS=Homo sapiens OX=9606 GN=SIAE PE=1 SV=1 |
| Q5RFU0 | 2.31e-33 | 873 | 1267 | 25 | 425 | Sialate O-acetylesterase OS=Pongo abelii OX=9601 GN=SIAE PE=2 SV=1 |
| P49008 | 4.70e-32 | 25 | 284 | 36 | 335 | Beta-hexosaminidase OS=Porphyromonas gingivalis (strain ATCC BAA-308 / W83) OX=242619 GN=nahA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
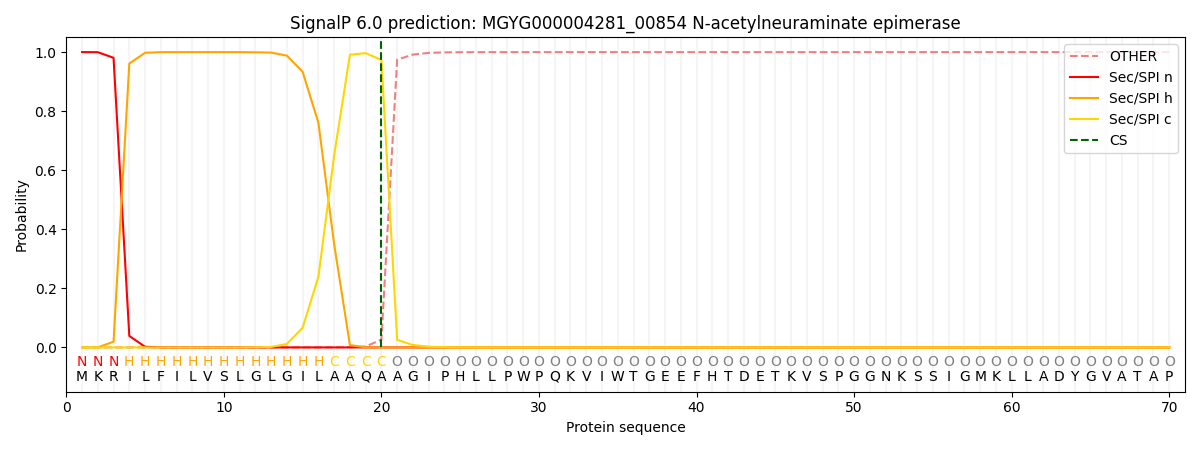
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000238 | 0.999087 | 0.000182 | 0.000160 | 0.000159 | 0.000149 |