You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004272_01442
You are here: Home > Sequence: MGYG000004272_01442
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
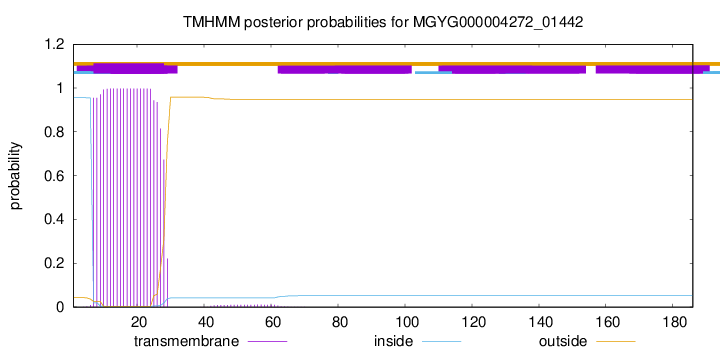
TMHMM annotations
Basic Information help
| Species | UMGS915 sp900546795 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Megasphaeraceae; UMGS915; UMGS915 sp900546795 | |||||||||||
| CAZyme ID | MGYG000004272_01442 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Soluble lytic murein transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8432; End: 8992 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 52 | 177 | 3.4e-30 | 0.7851851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16896 | LT_Slt70-like | 2.23e-74 | 35 | 170 | 1 | 137 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd13401 | Slt70-like | 1.67e-57 | 33 | 170 | 1 | 139 | 70kDa soluble lytic transglycosylase (Slt70) and similar proteins. Catalytic domain of the 70kda soluble lytic transglycosylase (LT)-like proteins, which also have an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
| cd00254 | LT-like | 1.88e-39 | 53 | 170 | 1 | 103 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG0741 | MltE | 1.26e-35 | 20 | 185 | 121 | 288 | Soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) [Cell wall/membrane/envelope biogenesis]. |
| pfam01464 | SLT | 5.46e-32 | 42 | 151 | 1 | 108 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXB81730.1 | 7.05e-67 | 2 | 185 | 6 | 190 |
| AXL22000.1 | 2.53e-62 | 5 | 184 | 9 | 189 |
| AVO73570.1 | 2.95e-56 | 2 | 184 | 7 | 189 |
| AVO26251.1 | 2.95e-56 | 2 | 184 | 7 | 189 |
| ALG43012.1 | 2.95e-56 | 2 | 184 | 7 | 189 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FBT_A | 9.00e-26 | 25 | 185 | 436 | 593 | ChainA, Lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 5OHU_A | 9.33e-26 | 25 | 185 | 465 | 622 | TheX-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa [Pseudomonas aeruginosa] |
| 6FC4_A | 2.29e-25 | 25 | 185 | 437 | 594 | ChainA, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 6FCQ_A | 2.29e-25 | 25 | 185 | 436 | 593 | ChainA, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCR_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCS_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCU_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 1QSA_A | 8.24e-23 | 14 | 185 | 429 | 603 | CrystalStructure Of The 70 Kda Soluble Lytic Transglycosylase Slt70 From Escherichia Coli At 1.65 Angstroms Resolution [Escherichia coli],1QTE_A Crystal Structure Of The 70 Kda Soluble Lytic Transglycosylase Slt70 From Escherichia Coli At 1.90 A Resolution In Complex With A 1,6- Anhydromurotripeptide [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0AGC3 | 4.59e-22 | 14 | 185 | 456 | 630 | Soluble lytic murein transglycosylase OS=Escherichia coli (strain K12) OX=83333 GN=slt PE=1 SV=1 |
| P0AGC4 | 4.59e-22 | 14 | 185 | 456 | 630 | Soluble lytic murein transglycosylase OS=Escherichia coli O157:H7 OX=83334 GN=slt PE=3 SV=1 |
| P39434 | 8.50e-22 | 14 | 185 | 456 | 630 | Soluble lytic murein transglycosylase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=slt PE=3 SV=2 |
| O31608 | 2.19e-14 | 49 | 170 | 71 | 172 | Putative murein lytic transglycosylase YjbJ OS=Bacillus subtilis (strain 168) OX=224308 GN=yjbJ PE=3 SV=1 |
| O31976 | 4.02e-14 | 33 | 142 | 1417 | 1518 | SPbeta prophage-derived uncharacterized transglycosylase YomI OS=Bacillus subtilis (strain 168) OX=224308 GN=yomI PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.988078 | 0.003198 | 0.008427 | 0.000012 | 0.000008 | 0.000318 |