You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004246_01924
You are here: Home > Sequence: MGYG000004246_01924
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM12669 sp900761935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; HGM12669; HGM12669 sp900761935 | |||||||||||
| CAZyme ID | MGYG000004246_01924 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10286; End: 13900 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT84 | 969 | 1180 | 3e-60 | 0.986046511627907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2943 | MdoH | 1.25e-07 | 551 | 773 | 347 | 602 | Membrane glycosyltransferase [Cell wall/membrane/envelope biogenesis, Carbohydrate transport and metabolism]. |
| pfam10091 | Glycoamylase | 3.43e-07 | 968 | 1167 | 3 | 203 | Putative glucoamylase. The structure of UniProt:Q5LIB7 has an alpha/alpha toroid fold and is similar structurally to a number of glucoamylases. Most of these structural homologs are glucoamylases, involved in breaking down complex sugars (e.g. starch). The biologically relevant state is likely to be monomeric. The putative active site is located at the centre of the toroid with a well defined large cavity. |
| PRK05454 | PRK05454 | 2.85e-06 | 551 | 706 | 327 | 523 | glucans biosynthesis glucosyltransferase MdoH. |
| cd04191 | Glucan_BSP_MdoH | 0.001 | 551 | 596 | 202 | 253 | Glucan_BSP_MdoH catalyzes the elongation of beta-1,2 polyglucose chains of glucan. Periplasmic Glucan Biosynthesis protein MdoH is a glucosyltransferase that catalyzes the elongation of beta-1,2 polyglucose chains of glucan, requiring a beta-glucoside as a primer and UDP-glucose as a substrate. Glucans are composed of 5 to 10 units of glucose forming a highly branched structure, where beta-1,2-linked glucose constitutes a linear backbone to which branches are attached by beta-1,6 linkages. In Escherichia coli, glucans are located in the periplasmic space, functioning as regulator of osmolarity. It is synthesized at a maximum when cells are grown in a medium with low osmolarity. It has been shown to span the cytoplasmic membrane. |
| pfam11329 | DUF3131 | 0.002 | 802 | 920 | 3 | 122 | Protein of unknown function (DUF3131). This bacterial family of proteins has no known function. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SHD75935.1 | 1.63e-152 | 136 | 1194 | 257 | 1362 |
| CDZ23197.1 | 3.49e-152 | 36 | 1194 | 69 | 1246 |
| ADU26013.1 | 3.94e-152 | 36 | 1194 | 65 | 1243 |
| AVQ95160.1 | 4.04e-152 | 36 | 1194 | 75 | 1253 |
| AYF37850.1 | 4.04e-152 | 36 | 1194 | 75 | 1253 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 1.43e-108 | 293 | 1198 | 480 | 1526 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000036 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
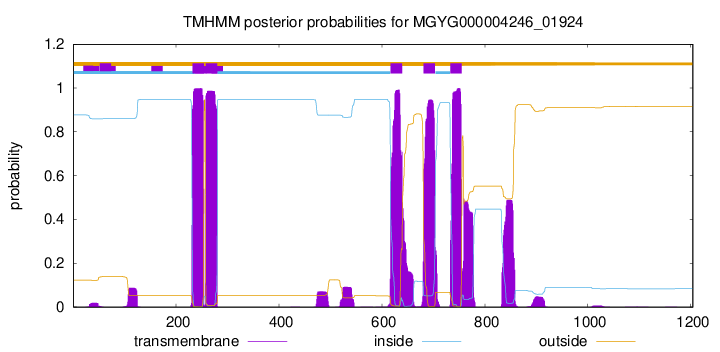
| start | end |
|---|---|
| 232 | 254 |
| 258 | 280 |
| 617 | 639 |
| 681 | 703 |
| 733 | 755 |
