You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004022_00317
You are here: Home > Sequence: MGYG000004022_00317
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
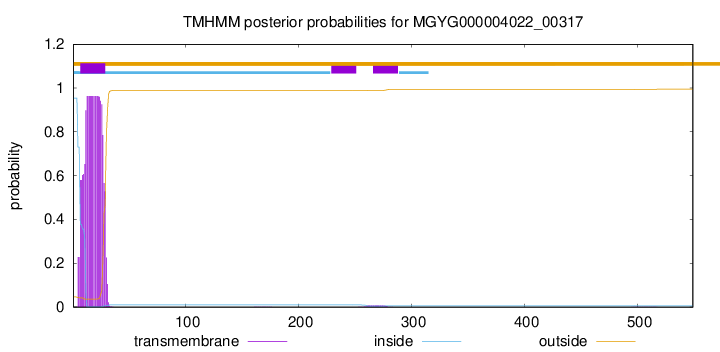
TMHMM annotations
Basic Information help
| Species | HGM13222 sp900757485 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; HGM13222; HGM13222 sp900757485 | |||||||||||
| CAZyme ID | MGYG000004022_00317 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 311429; End: 313078 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 273 | 530 | 5.4e-31 | 0.7446153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 7.90e-09 | 311 | 466 | 247 | 397 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN02793 | PLN02793 | 4.34e-05 | 275 | 381 | 138 | 259 | Probable polygalacturonase |
| pfam00295 | Glyco_hydro_28 | 0.001 | 303 | 549 | 86 | 304 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU07174.1 | 3.79e-179 | 107 | 548 | 49 | 489 |
| QTH43708.1 | 4.75e-48 | 104 | 497 | 2 | 414 |
| AXC14441.1 | 7.72e-44 | 150 | 548 | 86 | 477 |
| SDS06655.1 | 9.11e-44 | 109 | 495 | 42 | 436 |
| APA66227.1 | 1.91e-43 | 109 | 495 | 29 | 423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1WMR_A | 9.45e-09 | 133 | 458 | 56 | 431 | CrystalStructure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1WMR_B Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_A Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_B Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],2Z8G_A Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger],2Z8G_B Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger] |
| 3WWG_A | 9.45e-09 | 133 | 458 | 56 | 431 | Crystalstructure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_B Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_C Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_D Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O00105 | 5.27e-08 | 133 | 458 | 71 | 446 | Isopullulanase OS=Aspergillus niger OX=5061 GN=ipuA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000103 | 0.003104 | 0.996814 | 0.000002 | 0.000003 | 0.000002 |