You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003957_00794
You are here: Home > Sequence: MGYG000003957_00794
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp014287585 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp014287585 | |||||||||||
| CAZyme ID | MGYG000003957_00794 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42581; End: 44131 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 81 | 466 | 2.7e-45 | 0.9636963696369637 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.28e-32 | 164 | 466 | 19 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.62e-29 | 164 | 466 | 62 | 308 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.08e-25 | 158 | 466 | 79 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA28189.1 | 1.92e-218 | 8 | 512 | 1 | 502 |
| QQZ02681.1 | 2.18e-203 | 29 | 515 | 19 | 494 |
| AWI10666.1 | 4.41e-200 | 35 | 516 | 1 | 480 |
| AVM47074.1 | 7.02e-196 | 24 | 516 | 7 | 496 |
| AHF92621.1 | 3.52e-192 | 28 | 506 | 11 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 8.03e-14 | 55 | 512 | 52 | 410 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 5.92e-13 | 55 | 512 | 52 | 410 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 1NQ6_A | 1.96e-12 | 164 | 439 | 64 | 272 | CrystalStructure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
| 3RDK_A | 1.56e-11 | 164 | 466 | 65 | 335 | Proteincrystal structure of xylanase A1 of Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RDK_B Protein crystal structure of xylanase A1 of Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_A Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_B Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_C Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_D Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_E Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_F Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_G Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_H Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],4E4P_A Second native structure of Xylanase A1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],4E4P_B Second native structure of Xylanase A1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2] |
| 7CPK_A | 5.56e-11 | 158 | 357 | 65 | 229 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q12603 | 1.63e-10 | 152 | 466 | 70 | 349 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
| C6CRV0 | 2.67e-10 | 164 | 466 | 578 | 848 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P07528 | 3.62e-10 | 158 | 357 | 109 | 273 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| O69231 | 6.10e-10 | 164 | 466 | 65 | 328 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
| O69230 | 9.80e-07 | 164 | 511 | 431 | 739 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
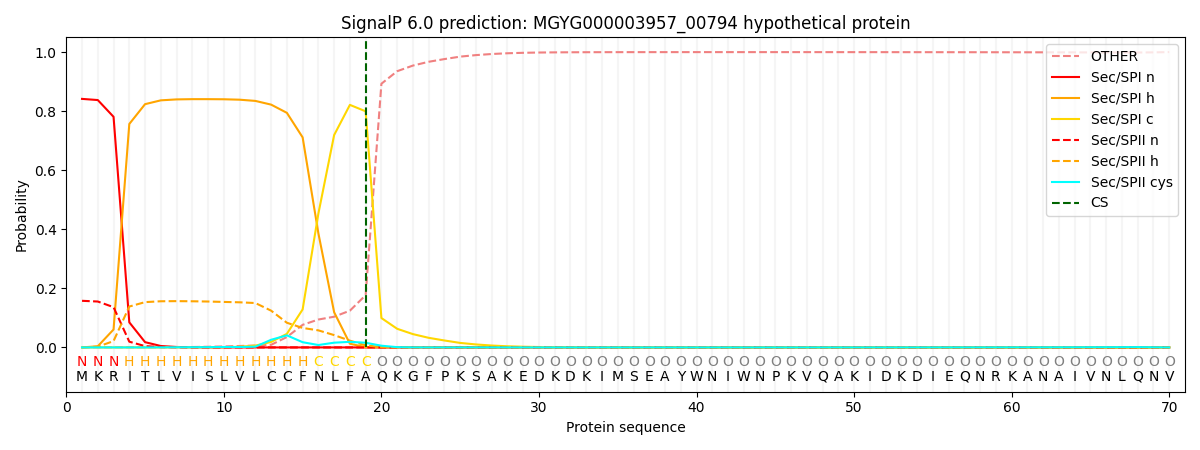
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001668 | 0.833200 | 0.164318 | 0.000271 | 0.000259 | 0.000259 |
