You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003911_00155
You are here: Home > Sequence: MGYG000003911_00155
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
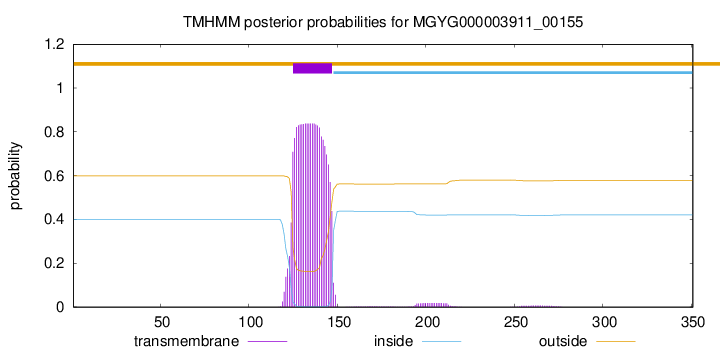
TMHMM annotations
Basic Information help
| Species | QAMM01 sp900762715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; QAMM01; QAMM01 sp900762715 | |||||||||||
| CAZyme ID | MGYG000003911_00155 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13879; End: 14934 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd07989 | LPLAT_AGPAT-like | 2.30e-28 | 139 | 319 | 4 | 183 | Lysophospholipid Acyltransferases (LPLATs) of Glycerophospholipid Biosynthesis: AGPAT-like. Lysophospholipid acyltransferase (LPLAT) superfamily member: acyltransferases of de novo and remodeling pathways of glycerophospholipid biosynthesis which catalyze the incorporation of an acyl group from either acylCoAs or acyl-acyl carrier proteins (acylACPs) into acceptors such as glycerol 3-phosphate, dihydroxyacetone phosphate or lyso-phosphatidic acid. Included in this subgroup are such LPLATs as 1-acyl-sn-glycerol-3-phosphate acyltransferase (AGPAT, PlsC), Tafazzin (product of Barth syndrome gene), and similar proteins. |
| COG0204 | PlsC | 2.68e-17 | 114 | 297 | 21 | 197 | 1-acyl-sn-glycerol-3-phosphate acyltransferase [Lipid transport and metabolism]. |
| smart00563 | PlsC | 5.66e-17 | 162 | 276 | 1 | 117 | Phosphate acyltransferases. Function in phospholipid biosynthesis and have either glycerolphosphate, 1-acylglycerolphosphate, or 2-acylglycerolphosphoethanolamine acyltransferase activities. Tafazzin, the product of the gene mutated in patients with Barth syndrome, is a member of this family. |
| pfam01553 | Acyltransferase | 8.16e-17 | 147 | 274 | 2 | 130 | Acyltransferase. This family contains acyltransferases involved in phospholipid biosynthesis and other proteins of unknown function. This family also includes tafazzin, the Barth syndrome gene. |
| PRK08633 | PRK08633 | 1.45e-09 | 132 | 315 | 414 | 602 | 2-acyl-glycerophospho-ethanolamine acyltransferase; Validated |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANW99047.1 | 1.20e-109 | 1 | 323 | 288 | 609 |
| ANX01575.1 | 1.20e-109 | 1 | 323 | 288 | 609 |
| AGC68714.1 | 1.20e-109 | 1 | 323 | 288 | 609 |
| AGI39723.1 | 1.20e-109 | 1 | 323 | 288 | 609 |
| AEJ20378.1 | 6.82e-58 | 1 | 323 | 309 | 623 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QHP_A | 8.84e-14 | 1 | 61 | 92 | 152 | Crystalstructure of the catalytic domain of cholesterol-alpha-glucosyltransferase from Helicobacter pylori [Helicobacter pylori 26695],3QHP_B Crystal structure of the catalytic domain of cholesterol-alpha-glucosyltransferase from Helicobacter pylori [Helicobacter pylori 26695] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GXU8 | 1.88e-06 | 144 | 274 | 178 | 306 | 1-acyl-sn-glycerol-3-phosphate acyltransferase LPAT1, chloroplastic OS=Arabidopsis thaliana OX=3702 GN=LPAT1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
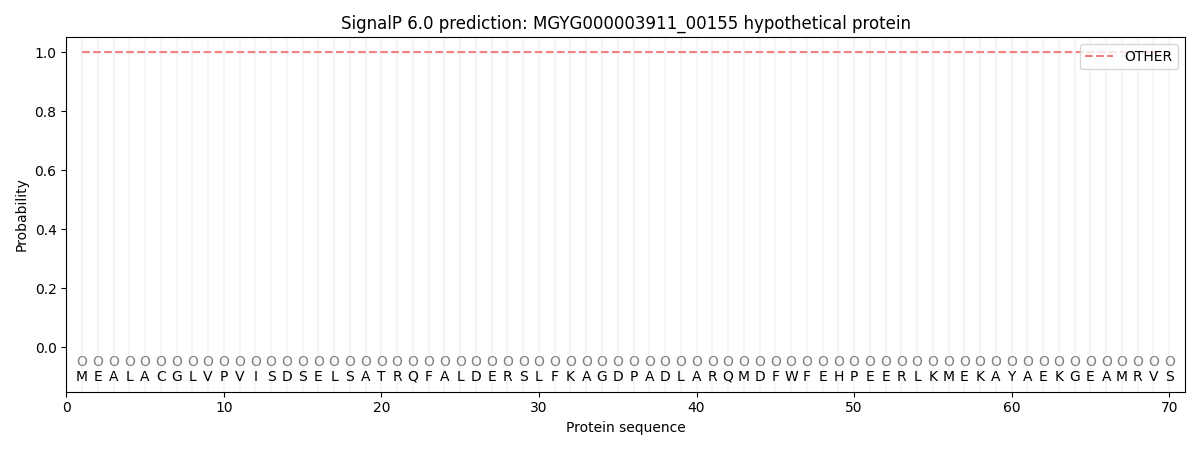
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000064 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |