You are browsing environment: HUMAN GUT
MGYG000003833_01255
Basic Information
help
Species
Lineage
Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; WRGP01;
CAZyme ID
MGYG000003833_01255
CAZy Family
GH163
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003833
2358901
MAG
United States
North America
Gene Location
Start: 512;
End: 2245
Strand: -
No EC number prediction in MGYG000003833_01255.
Family
Start
End
Evalue
family coverage
GH163
173
452
1.6e-86
0.9960159362549801
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam16126
DUF4838
9.37e-101
166
452
1
263
Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown.
more
pfam02838
Glyco_hydro_20b
8.82e-06
12
88
26
101
Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold.
more
NF033446
BREX_PglZ_2
4.07e-05
417
571
224
379
BREX-2 system phosphatase PglZ.
more
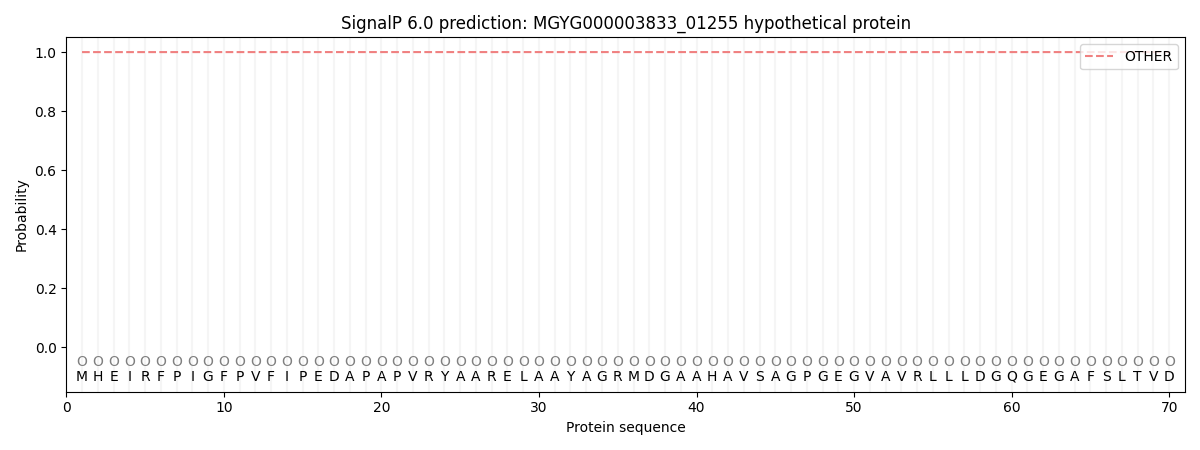
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000055
0.000005
0.000000
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000003833_01255.