You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003806_01875
You are here: Home > Sequence: MGYG000003806_01875
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
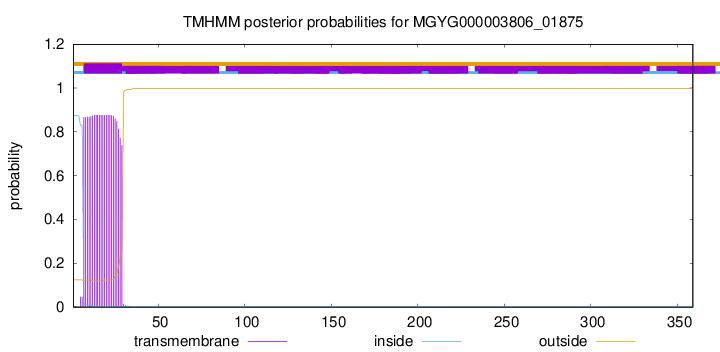
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; | |||||||||||
| CAZyme ID | MGYG000003806_01875 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | Diacylglycerol acyltransferase/mycolyltransferase Ag85A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7135; End: 8214 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 73 | 353 | 4.4e-45 | 0.9779735682819384 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0627 | FrmB | 8.30e-52 | 71 | 358 | 45 | 313 | S-formylglutathione hydrolase FrmB [Defense mechanisms]. |
| pfam00756 | Esterase | 4.71e-17 | 73 | 347 | 1 | 240 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVI99079.1 | 2.07e-127 | 8 | 358 | 7 | 354 |
| AOX06778.1 | 2.94e-120 | 1 | 352 | 1 | 370 |
| AKK10240.1 | 6.25e-105 | 12 | 354 | 11 | 366 |
| AMY54059.1 | 6.51e-75 | 75 | 356 | 62 | 338 |
| QLY34586.1 | 1.06e-74 | 78 | 356 | 2 | 274 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4H18_A | 5.19e-118 | 1 | 357 | 4 | 361 | Threedimensional structure of corynomycoloyl tranferase C [Corynebacterium glutamicum ATCC 13032],4H18_B Three dimensional structure of corynomycoloyl tranferase C [Corynebacterium glutamicum ATCC 13032],4H18_C Three dimensional structure of corynomycoloyl tranferase C [Corynebacterium glutamicum ATCC 13032],4H18_D Three dimensional structure of corynomycoloyl tranferase C [Corynebacterium glutamicum ATCC 13032] |
| 6SX4_AAA | 1.53e-28 | 90 | 356 | 92 | 342 | ChainAAA, Protein PS1 [Corynebacterium glutamicum],6SX4_BBB Chain BBB, Protein PS1 [Corynebacterium glutamicum],6SX4_CCC Chain CCC, Protein PS1 [Corynebacterium glutamicum],6SX4_DDD Chain DDD, Protein PS1 [Corynebacterium glutamicum] |
| 1SFR_A | 1.08e-22 | 69 | 359 | 12 | 285 | ChainA, Antigen 85-A [Mycobacterium tuberculosis],1SFR_B Chain B, Antigen 85-A [Mycobacterium tuberculosis],1SFR_C Chain C, Antigen 85-A [Mycobacterium tuberculosis] |
| 4MQL_A | 9.18e-21 | 96 | 358 | 36 | 283 | ChainA, Diacylglycerol acyltransferase/mycolyltransferase Ag85C [Mycobacterium tuberculosis] |
| 7MYG_A | 2.17e-20 | 96 | 358 | 30 | 277 | ChainA, Diacylglycerol acyltransferase [Mycobacterium tuberculosis],7MYG_B Chain B, Diacylglycerol acyltransferase [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1D7 | 1.81e-31 | 34 | 356 | 78 | 385 | Protein PS1 OS=Corynebacterium melassecola OX=41643 GN=csp1 PE=4 SV=1 |
| P0C1D6 | 2.85e-30 | 34 | 356 | 78 | 385 | Protein PS1 OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=csp1 PE=1 SV=1 |
| P0C2T1 | 9.77e-22 | 69 | 359 | 54 | 327 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=fbpA PE=3 SV=1 |
| P9WQP3 | 9.77e-22 | 69 | 359 | 54 | 327 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=fbpA PE=1 SV=1 |
| P9WQP2 | 9.77e-22 | 69 | 359 | 54 | 327 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=fbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000515 | 0.998454 | 0.000200 | 0.000353 | 0.000252 | 0.000199 |