You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003761_01945
You are here: Home > Sequence: MGYG000003761_01945
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas maltophilia_S | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia_S | |||||||||||
| CAZyme ID | MGYG000003761_01945 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3333; End: 5042 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 4 | 535 | 8e-98 | 0.9740740740740741 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.31e-44 | 11 | 458 | 11 | 448 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 2.92e-14 | 63 | 226 | 1 | 158 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279 | arnT | 1.64e-07 | 28 | 330 | 26 | 320 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam02366 | PMT | 2.22e-07 | 88 | 227 | 91 | 233 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This is a family of Dolichyl-phosphate-mannose-protein mannosyltransferase proteins EC:2.4.1.109. These proteins are responsible for O-linked glycosylation of proteins, they catalyze the reaction:- Dolichyl phosphate D-mannose + protein <=> dolichyl phosphate + O-D-mannosyl-protein. Also in this family is Drosophila rotated abdomen protein which is a putative mannosyltransferase. This family appears to be distantly related to pfam02516 (A Bateman pers. obs.). This family also contains sequences from ArnTs (4-amino-4-deoxy-L-arabinose lipid A transferase). They catalyze the addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) to the lipid A moiety of the lipopolysaccharide. This is a critical modification enabling bacteria (e.g. Escherichia coli and Salmonella typhimurium) to resist killing by antimicrobial peptides such as polymyxins. Members such as undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase are predicted to have 12 trans-membrane regions. The N-terminal portion of these proteins is hypothesized to have a conserved glycosylation activity which is shared between distantly related oligosaccharyltransferases ArnT and PglB families. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDL29643.1 | 0.0 | 1 | 569 | 1 | 569 |
| AEM52938.1 | 0.0 | 1 | 569 | 1 | 569 |
| AUI09165.1 | 0.0 | 1 | 569 | 1 | 569 |
| QGL82024.1 | 0.0 | 1 | 569 | 1 | 569 |
| QNG76753.1 | 0.0 | 1 | 569 | 1 | 569 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 3.42e-09 | 4 | 467 | 28 | 483 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6DAW3 | 3.10e-17 | 8 | 377 | 9 | 369 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium carotovorum subsp. carotovorum (strain PC1) OX=561230 GN=arnT PE=3 SV=1 |
| Q6D2F3 | 6.96e-16 | 3 | 346 | 3 | 338 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=arnT PE=3 SV=1 |
| A8FRR0 | 1.51e-14 | 12 | 340 | 14 | 329 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1 |
| B4ETL9 | 8.28e-14 | 1 | 410 | 1 | 397 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Proteus mirabilis (strain HI4320) OX=529507 GN=arnT2 PE=3 SV=1 |
| Q4ZSZ0 | 7.77e-13 | 8 | 446 | 7 | 425 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999611 | 0.000182 | 0.000025 | 0.000001 | 0.000001 | 0.000230 |
TMHMM Annotations download full data without filtering help
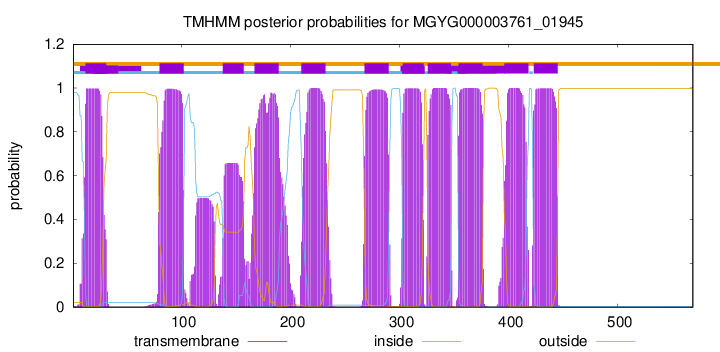
| start | end |
|---|---|
| 12 | 31 |
| 80 | 102 |
| 138 | 157 |
| 167 | 189 |
| 210 | 232 |
| 268 | 290 |
| 303 | 322 |
| 326 | 347 |
| 354 | 376 |
| 396 | 418 |
| 423 | 445 |
