You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003533_02005
You are here: Home > Sequence: MGYG000003533_02005
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | SFTJ01 sp900769345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; SFTJ01; SFTJ01 sp900769345 | |||||||||||
| CAZyme ID | MGYG000003533_02005 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10544; End: 11314 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 35 | 236 | 9.7e-28 | 0.8810572687224669 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 6.41e-44 | 32 | 255 | 171 | 387 | Predicted peptidase [General function prediction only]. |
| COG1506 | DAP2 | 2.16e-14 | 30 | 235 | 373 | 595 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG3509 | LpqC | 1.45e-12 | 1 | 226 | 8 | 230 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG0400 | YpfH | 1.45e-11 | 46 | 231 | 13 | 186 | Predicted esterase [General function prediction only]. |
| pfam02230 | Abhydrolase_2 | 4.63e-10 | 52 | 238 | 15 | 203 | Phospholipase/Carboxylesterase. This family consists of both phospholipases and carboxylesterases with broad substrate specificity, and is structurally related to alpha/beta hydrolases pfam00561. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61582.1 | 6.27e-41 | 37 | 254 | 827 | 1042 |
| ABS60377.1 | 3.40e-40 | 44 | 255 | 30 | 245 |
| QDU56037.1 | 1.74e-33 | 49 | 255 | 817 | 1007 |
| ACR12533.1 | 5.35e-24 | 34 | 239 | 63 | 270 |
| QEY17854.1 | 5.74e-23 | 20 | 255 | 781 | 1041 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 3.73e-45 | 32 | 255 | 155 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 1.02e-23 | 28 | 255 | 14 | 217 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 4Q82_A | 4.56e-11 | 49 | 255 | 78 | 277 | CrystalStructure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365],4Q82_B Crystal Structure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0J968 | 1.49e-07 | 51 | 234 | 9 | 199 | Probable carboxylesterase Os04g0669600 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0669600 PE=2 SV=1 |
| P52090 | 3.92e-07 | 52 | 217 | 66 | 247 | Poly(3-hydroxyalkanoate) depolymerase C OS=Paucimonas lemoignei OX=29443 GN=phaZ1 PE=3 SV=1 |
| Q9HGR3 | 7.59e-06 | 46 | 181 | 43 | 164 | Feruloyl esterase B OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=fae-1 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
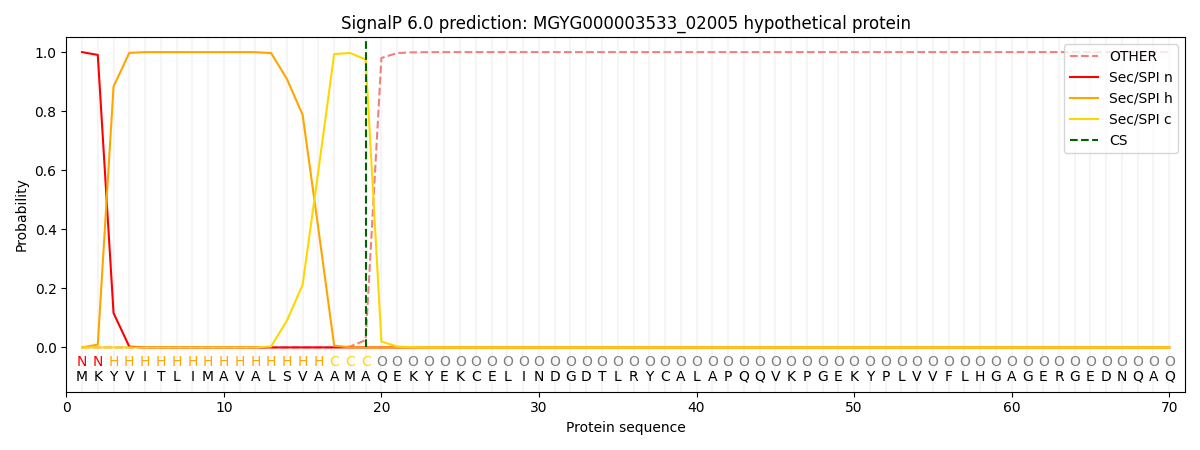
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000218 | 0.999193 | 0.000150 | 0.000142 | 0.000135 | 0.000129 |
