You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003528_02314
You are here: Home > Sequence: MGYG000003528_02314
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAAGGB01 sp900769285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; UBA1407; UBA1407; CAAGGB01; CAAGGB01 sp900769285 | |||||||||||
| CAZyme ID | MGYG000003528_02314 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11843; End: 13459 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 190 | 456 | 1e-44 | 0.7381818181818182 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.57e-26 | 182 | 451 | 14 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 9.31e-14 | 189 | 476 | 71 | 383 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam05448 | AXE1 | 6.16e-10 | 449 | 524 | 234 | 307 | Acetyl xylan esterase (AXE1). This family consists of several bacterial acetyl xylan esterase proteins. Acetyl xylan esterases are enzymes that hydrolyze the ester linkages of the acetyl groups in position 2 and/or 3 of the xylose moieties of natural acetylated xylan from hardwood. These enzymes are one of the accessory enzymes which are part of the xylanolytic system, together with xylanases, beta-xylosidases, alpha-arabinofuranosidases and methylglucuronidases; these are all required for the complete hydrolysis of xylan. |
| COG3458 | Axe1 | 8.66e-08 | 450 | 535 | 234 | 310 | Cephalosporin-C deacetylase or related acetyl esterase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM45191.1 | 8.13e-127 | 31 | 462 | 43 | 476 |
| AVM45735.1 | 5.36e-101 | 31 | 460 | 33 | 479 |
| AVM44368.1 | 2.07e-93 | 7 | 463 | 22 | 495 |
| AVM43793.1 | 6.09e-88 | 1 | 463 | 1 | 480 |
| AVM47005.1 | 3.44e-77 | 8 | 460 | 74 | 542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W0K_A | 1.33e-19 | 225 | 460 | 45 | 292 | CrystalStructure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus],3W0K_B Crystal Structure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus] |
| 1CEC_A | 4.67e-16 | 230 | 473 | 65 | 327 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
| 7EC9_A | 8.24e-16 | 193 | 442 | 38 | 295 | ChainA, Endoglucanase [Thermotoga maritima MSB8],7EC9_B Chain B, Endoglucanase [Thermotoga maritima MSB8],7EFZ_A Chain A, Endoglucanase [Thermotoga maritima MSB8],7EFZ_B Chain B, Endoglucanase [Thermotoga maritima MSB8] |
| 1CEN_A | 1.13e-15 | 230 | 473 | 65 | 327 | ChainA, CELLULASE CELC [Acetivibrio thermocellus],1CEO_A Chain A, CELLULASE CELC [Acetivibrio thermocellus] |
| 1VJZ_A | 2.67e-15 | 193 | 442 | 38 | 295 | Crystalstructure of Endoglucanase (TM1752) from Thermotoga maritima at 2.05 A resolution [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16169 | 1.32e-15 | 191 | 445 | 30 | 285 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
| A3DJ77 | 1.90e-15 | 230 | 473 | 65 | 327 | Endoglucanase C OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celC PE=3 SV=1 |
| P23340 | 1.90e-15 | 230 | 473 | 65 | 327 | Endoglucanase C307 OS=Clostridium sp. (strain F1) OX=1508 GN=celC307 PE=1 SV=1 |
| P0C2S3 | 2.56e-15 | 230 | 473 | 65 | 327 | Endoglucanase C OS=Acetivibrio thermocellus OX=1515 GN=celC PE=1 SV=1 |
| P54583 | 9.89e-14 | 236 | 461 | 138 | 364 | Endoglucanase E1 OS=Acidothermus cellulolyticus (strain ATCC 43068 / DSM 8971 / 11B) OX=351607 GN=Acel_0614 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
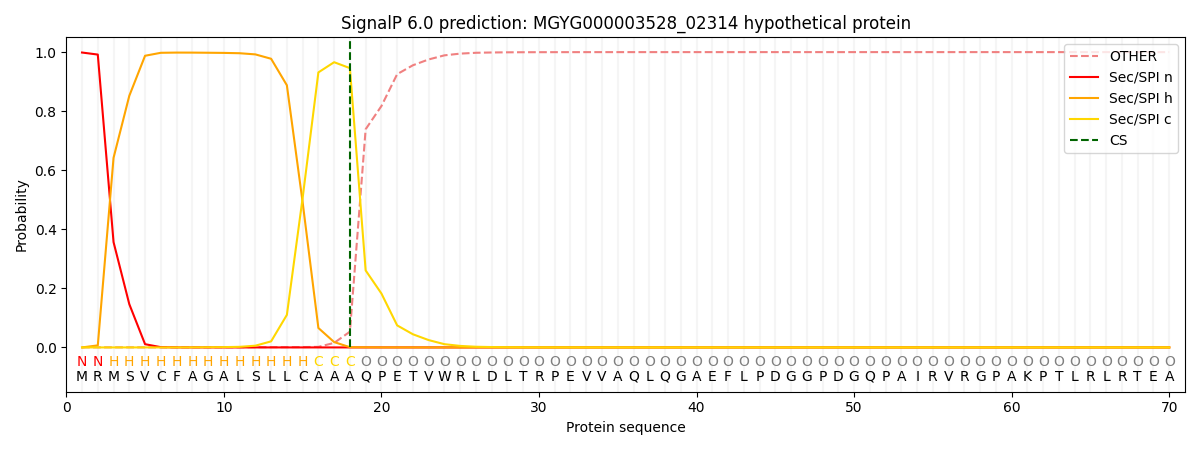
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001196 | 0.996428 | 0.001683 | 0.000240 | 0.000226 | 0.000212 |
