You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003525_00021
You are here: Home > Sequence: MGYG000003525_00021
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp004556005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp004556005 | |||||||||||
| CAZyme ID | MGYG000003525_00021 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28235; End: 30751 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 65 | 379 | 9.3e-95 | 0.9930795847750865 |
| CE7 | 548 | 823 | 6.6e-78 | 0.9169329073482428 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05448 | AXE1 | 5.61e-65 | 552 | 817 | 21 | 299 | Acetyl xylan esterase (AXE1). This family consists of several bacterial acetyl xylan esterase proteins. Acetyl xylan esterases are enzymes that hydrolyze the ester linkages of the acetyl groups in position 2 and/or 3 of the xylose moieties of natural acetylated xylan from hardwood. These enzymes are one of the accessory enzymes which are part of the xylanolytic system, together with xylanases, beta-xylosidases, alpha-arabinofuranosidases and methylglucuronidases; these are all required for the complete hydrolysis of xylan. |
| COG3458 | Axe1 | 1.58e-59 | 543 | 817 | 13 | 299 | Cephalosporin-C deacetylase or related acetyl esterase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG3934 | COG3934 | 1.12e-33 | 32 | 440 | 49 | 476 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| COG1506 | DAP2 | 2.25e-08 | 589 | 794 | 367 | 568 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam00326 | Peptidase_S9 | 8.45e-06 | 676 | 793 | 45 | 160 | Prolyl oligopeptidase family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SJX74200.1 | 1.27e-201 | 305 | 834 | 2 | 525 |
| QQA30547.1 | 2.72e-170 | 6 | 423 | 6 | 422 |
| ADD61406.1 | 2.72e-170 | 6 | 423 | 6 | 422 |
| ADD62011.1 | 2.72e-170 | 6 | 423 | 6 | 422 |
| QUT33697.1 | 2.72e-170 | 6 | 423 | 6 | 422 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1UUQ_A | 4.81e-90 | 19 | 424 | 16 | 424 | Exo-mannosidasefrom Cellvibrio mixtus [Cellvibrio mixtus],1UZ4_A Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis [Cellvibrio mixtus],7ODJ_AAA Chain AAA, Man5A [Cellvibrio mixtus] |
| 4LYP_A | 5.39e-65 | 22 | 424 | 32 | 435 | CrystalStructure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei],4LYP_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei] |
| 4LYQ_A | 3.76e-64 | 22 | 424 | 32 | 435 | CrystalStructure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant [Rhizomucor miehei],4NRR_A Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose [Rhizomucor miehei],4NRR_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose [Rhizomucor miehei],4NRS_A Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose [Rhizomucor miehei],4NRS_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose [Rhizomucor miehei] |
| 4LYR_A | 3.76e-64 | 22 | 424 | 32 | 435 | GlycosideHydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant [Rhizomucor miehei] |
| 1L7A_A | 8.57e-47 | 552 | 836 | 22 | 315 | structuralGenomics, crystal structure of Cephalosporin C deacetylase [Bacillus subtilis],1L7A_B structural Genomics, crystal structure of Cephalosporin C deacetylase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94388 | 1.20e-45 | 552 | 834 | 22 | 313 | Cephalosporin-C deacetylase OS=Bacillus subtilis (strain 168) OX=224308 GN=cah PE=1 SV=1 |
| Q0JKM9 | 3.30e-42 | 27 | 377 | 37 | 371 | Mannan endo-1,4-beta-mannosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN1 PE=2 SV=2 |
| Q9FZ29 | 5.14e-38 | 6 | 377 | 4 | 361 | Mannan endo-1,4-beta-mannosidase 1 OS=Arabidopsis thaliana OX=3702 GN=MAN1 PE=2 SV=1 |
| Q9WXT2 | 5.82e-37 | 552 | 817 | 22 | 303 | Cephalosporin-C deacetylase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=axeA PE=1 SV=1 |
| Q5W6G0 | 6.96e-37 | 102 | 377 | 144 | 416 | Putative mannan endo-1,4-beta-mannosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN5 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
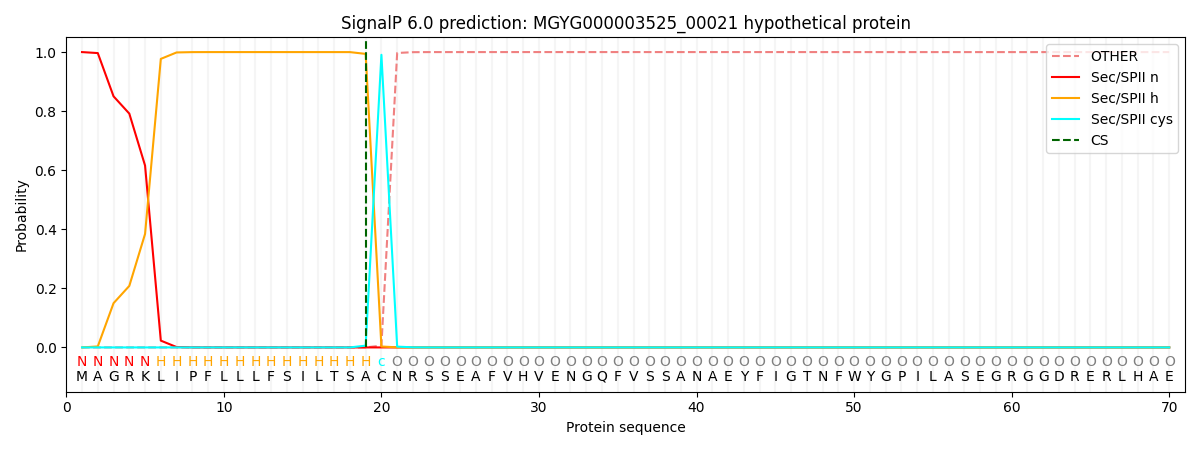
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000061 | 0.000000 | 0.000000 | 0.000000 |
