You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003520_01582
You are here: Home > Sequence: MGYG000003520_01582
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900769145 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900769145 | |||||||||||
| CAZyme ID | MGYG000003520_01582 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24462; End: 27566 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 57 | 913 | 1.4e-217 | 0.9667553191489362 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 27 | 1030 | 17 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 27 | 1031 | 4 | 998 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 8.73e-175 | 65 | 911 | 14 | 803 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 2.67e-101 | 360 | 652 | 5 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| smart01038 | Bgal_small_N | 3.97e-84 | 765 | 1030 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFL78169.1 | 0.0 | 1 | 1030 | 1 | 1071 |
| BBL00988.1 | 0.0 | 1 | 1030 | 1 | 1069 |
| BBL08913.1 | 0.0 | 1 | 1030 | 1 | 1069 |
| BBL11705.1 | 0.0 | 1 | 1030 | 1 | 1069 |
| EDY95440.1 | 0.0 | 27 | 1033 | 21 | 1072 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 9.03e-216 | 64 | 1028 | 39 | 977 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 9.29e-216 | 64 | 1028 | 40 | 978 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3MUY_1 | 2.12e-191 | 27 | 1025 | 16 | 1017 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
| 1F4A_A | 3.97e-191 | 27 | 1025 | 14 | 1015 | E.COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_B E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_C E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_D E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4H_A E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_B E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_C E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_D E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli] |
| 5A1A_A | 4.08e-191 | 27 | 1025 | 15 | 1016 | 2.2A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12],5A1A_B 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12],5A1A_C 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12],5A1A_D 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 5.09e-215 | 64 | 1028 | 40 | 978 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 4.67e-208 | 18 | 1010 | 11 | 1012 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| A5F5U6 | 3.60e-193 | 27 | 1026 | 13 | 1012 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| Q8D4H3 | 2.49e-192 | 27 | 1026 | 13 | 1023 | Beta-galactosidase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=lacZ PE=3 SV=2 |
| B7N8Q1 | 1.09e-191 | 27 | 1025 | 17 | 1018 | Beta-galactosidase OS=Escherichia coli O17:K52:H18 (strain UMN026 / ExPEC) OX=585056 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
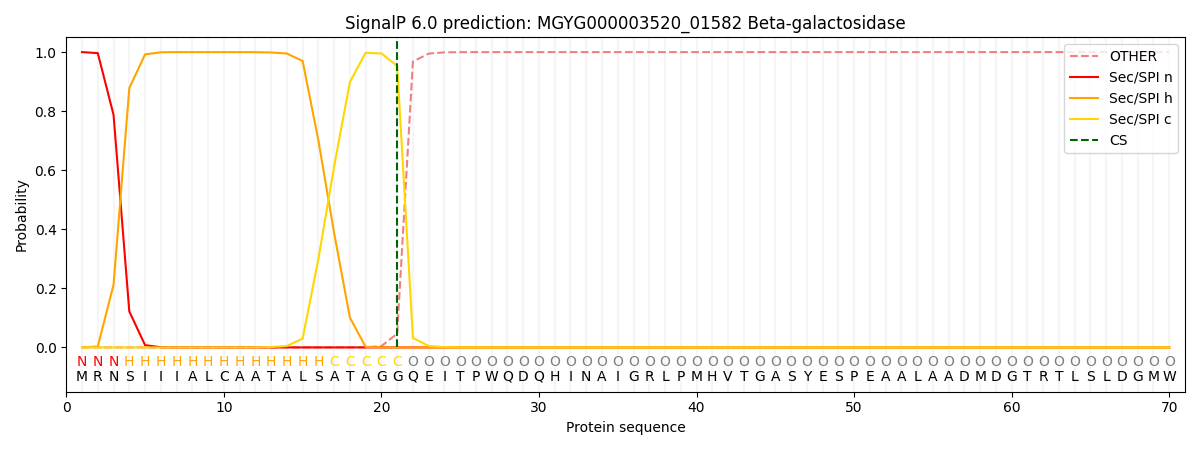
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000263 | 0.999028 | 0.000187 | 0.000191 | 0.000164 | 0.000145 |
