You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003401_01317
You are here: Home > Sequence: MGYG000003401_01317
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus capitis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus capitis | |||||||||||
| CAZyme ID | MGYG000003401_01317 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | N-acetylmuramoyl-L-alanine amidase sle1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 290492; End: 291502 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM50 | 90 | 131 | 1.5e-17 | 0.95 |
| CBM50 | 158 | 200 | 2e-16 | 0.975 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3942 | COG3942 | 2.70e-34 | 221 | 336 | 57 | 171 | Surface antigen [Cell wall/membrane/envelope biogenesis]. |
| PRK06347 | PRK06347 | 4.09e-22 | 31 | 199 | 335 | 523 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 3.26e-21 | 31 | 199 | 410 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK08581 | PRK08581 | 1.52e-18 | 232 | 325 | 508 | 606 | amidase domain-containing protein. |
| pfam01476 | LysM | 1.01e-13 | 90 | 130 | 1 | 41 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKL93276.1 | 1.28e-214 | 1 | 336 | 1 | 336 |
| QOX61610.1 | 1.28e-214 | 1 | 336 | 1 | 336 |
| BAW91733.1 | 1.28e-214 | 1 | 336 | 1 | 336 |
| QKH91955.1 | 2.12e-210 | 1 | 336 | 1 | 333 |
| ATN03628.1 | 3.07e-206 | 1 | 336 | 1 | 336 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2K3A_A | 2.34e-31 | 230 | 336 | 50 | 153 | ChainA, CHAP domain protein [Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 = NCTC 7292] |
| 2LRJ_A | 3.99e-30 | 230 | 336 | 9 | 112 | ChainA, Staphyloxanthin biosynthesis protein, putative [Staphylococcus aureus subsp. aureus COL] |
| 5T1Q_A | 2.54e-16 | 232 | 324 | 248 | 345 | ChainA, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_B Chain B, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_C Chain C, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_D Chain D, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 4UZ2_A | 7.20e-09 | 90 | 131 | 5 | 46 | Crystalstructure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_B Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_C Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_D Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ3_A Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8],4UZ3_B Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8],4UZ3_C Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8] |
| 4XCM_A | 4.34e-07 | 90 | 131 | 5 | 46 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CMN2 | 3.37e-185 | 1 | 336 | 1 | 324 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=sle1 PE=3 SV=1 |
| Q5HRU2 | 3.37e-185 | 1 | 336 | 1 | 324 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=sle1 PE=3 SV=1 |
| Q6GJK9 | 1.97e-163 | 1 | 336 | 1 | 334 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=sle1 PE=3 SV=1 |
| Q6GC24 | 2.79e-163 | 1 | 336 | 1 | 334 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=sle1 PE=3 SV=1 |
| Q5HIL2 | 2.79e-163 | 1 | 336 | 1 | 334 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain COL) OX=93062 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
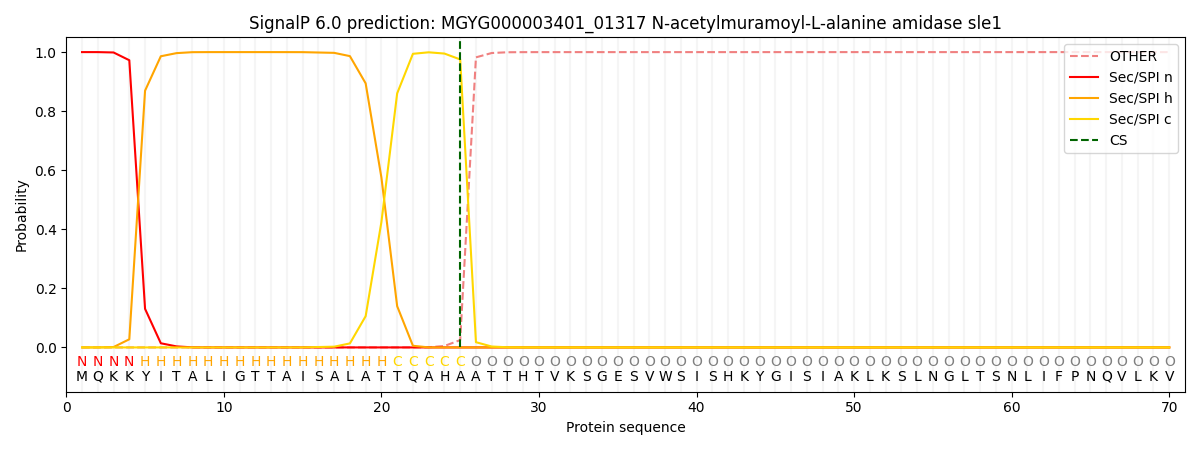
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000254 | 0.999005 | 0.000168 | 0.000209 | 0.000181 | 0.000153 |
