You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003355_00794
You are here: Home > Sequence: MGYG000003355_00794
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium phytofermentans_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; Lachnoclostridium phytofermentans_A | |||||||||||
| CAZyme ID | MGYG000003355_00794 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 46169; End: 53677 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 1876 | 2257 | 2e-129 | 0.9813333333333333 |
| CE8 | 601 | 966 | 2.8e-46 | 0.9166666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 8.84e-26 | 585 | 966 | 76 | 390 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02773 | PLN02773 | 3.56e-25 | 602 | 966 | 16 | 284 | pectinesterase |
| pfam01095 | Pectinesterase | 7.67e-22 | 601 | 984 | 10 | 298 | Pectinesterase. |
| PLN02682 | PLN02682 | 1.88e-18 | 602 | 987 | 81 | 366 | pectinesterase family protein |
| PLN02432 | PLN02432 | 7.67e-18 | 602 | 966 | 22 | 274 | putative pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX44216.1 | 0.0 | 1162 | 2502 | 81 | 1431 |
| ADL35206.1 | 0.0 | 30 | 2277 | 130 | 2463 |
| BCJ94249.1 | 0.0 | 1264 | 2290 | 178 | 1218 |
| AFK85374.1 | 1.10e-284 | 36 | 996 | 754 | 1730 |
| ACU08321.1 | 6.92e-239 | 28 | 989 | 14 | 989 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 3.07e-44 | 1881 | 2275 | 18 | 386 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 7.15e-20 | 1877 | 2141 | 2 | 303 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 5C1C_A | 7.98e-16 | 770 | 951 | 107 | 266 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 7.98e-16 | 594 | 951 | 11 | 266 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 2NSP_A | 4.98e-10 | 707 | 902 | 77 | 250 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 4.44e-56 | 1527 | 2163 | 85 | 646 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A6 | 5.83e-45 | 1881 | 2275 | 43 | 411 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 2.97e-43 | 1881 | 2275 | 43 | 411 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| Q4WBT5 | 9.29e-18 | 780 | 988 | 141 | 317 | Probable pectinesterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pmeA PE=3 SV=1 |
| B0Y9F9 | 9.29e-18 | 780 | 988 | 141 | 317 | Probable pectinesterase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pmeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
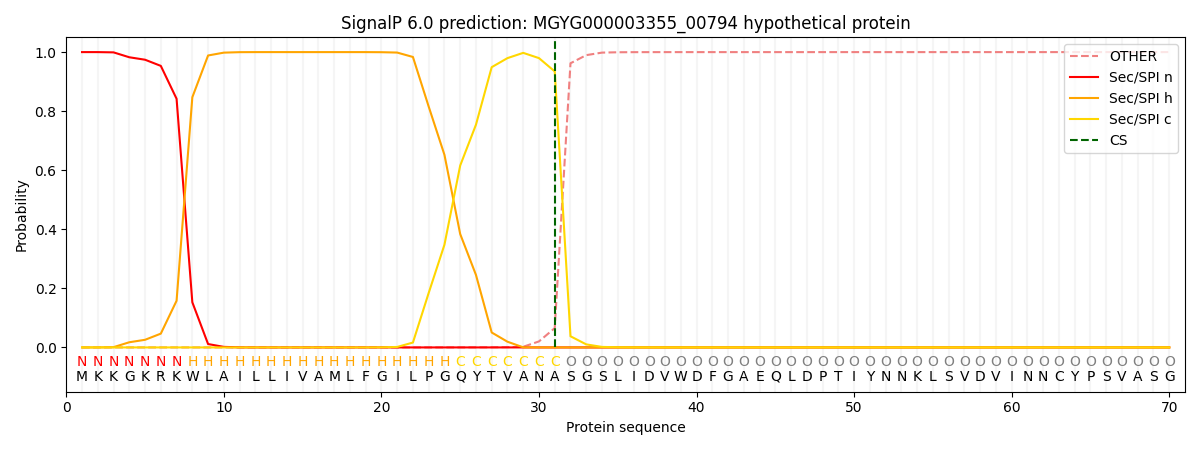
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000203 | 0.999209 | 0.000157 | 0.000159 | 0.000135 | 0.000129 |