You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003351_03708
You are here: Home > Sequence: MGYG000003351_03708
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900765785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900765785 | |||||||||||
| CAZyme ID | MGYG000003351_03708 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5335; End: 6897 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 56 | 510 | 1.1e-211 | 0.9977777777777778 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 1.05e-19 | 179 | 462 | 121 | 379 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02057 | Glyco_hydro_59 | 2.14e-08 | 162 | 372 | 78 | 252 | Glycosyl hydrolase family 59. |
| pfam14587 | Glyco_hydr_30_2 | 1.00e-06 | 157 | 384 | 103 | 366 | O-Glycosyl hydrolase family 30. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ39695.1 | 0.0 | 1 | 520 | 1 | 520 |
| QMW85450.1 | 0.0 | 1 | 520 | 1 | 520 |
| AAO78132.1 | 0.0 | 1 | 520 | 1 | 520 |
| QUT73128.1 | 0.0 | 1 | 520 | 1 | 520 |
| QQA08798.1 | 0.0 | 1 | 520 | 1 | 520 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7O0E_A | 5.09e-14 | 65 | 510 | 13 | 445 | ChainA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
| 5CXP_A | 1.19e-13 | 177 | 509 | 82 | 384 | X-raycrystallographic protein structure of the glycoside hydrolase family 30 subfamily 8 xylanase, Xyn30A, from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
| 7NCX_AAA | 1.32e-13 | 65 | 510 | 20 | 452 | ChainAAA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
| 3KE0_A | 1.85e-08 | 167 | 458 | 165 | 466 | ChainA, Glucosylceramidase [Homo sapiens],3KE0_B Chain B, Glucosylceramidase [Homo sapiens],3KEH_A Chain A, Glucocerebrosidase [Homo sapiens],3KEH_B Chain B, Glucocerebrosidase [Homo sapiens] |
| 2WNW_A | 2.23e-08 | 174 | 278 | 137 | 240 | Thecrystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],2WNW_B The crystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2Q1N4 | 5.53e-14 | 65 | 510 | 37 | 469 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
| P17439 | 1.94e-08 | 167 | 458 | 184 | 484 | Lysosomal acid glucosylceramidase OS=Mus musculus OX=10090 GN=Gba PE=1 SV=1 |
| P04062 | 1.41e-07 | 167 | 458 | 204 | 505 | Lysosomal acid glucosylceramidase OS=Homo sapiens OX=9606 GN=GBA PE=1 SV=3 |
| Q9BDT0 | 1.41e-07 | 167 | 458 | 204 | 505 | Lysosomal acid glucosylceramidase OS=Pan troglodytes OX=9598 GN=GBA PE=3 SV=1 |
| Q5R8E3 | 3.26e-07 | 167 | 458 | 204 | 505 | Lysosomal acid glucosylceramidase OS=Pongo abelii OX=9601 GN=GBA PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
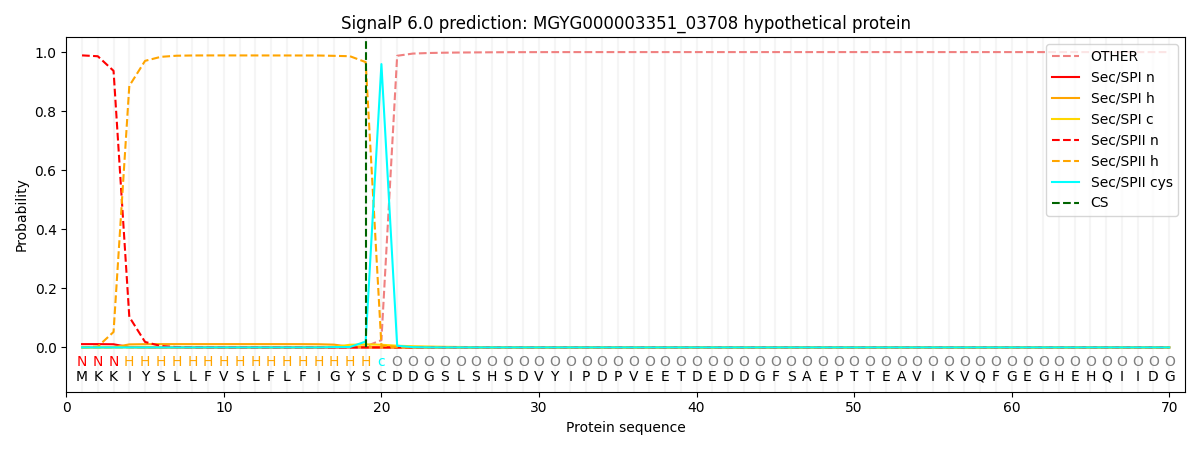
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000046 | 0.010857 | 0.989133 | 0.000005 | 0.000005 | 0.000005 |
