You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003252_03196
You are here: Home > Sequence: MGYG000003252_03196
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900761785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900761785 | |||||||||||
| CAZyme ID | MGYG000003252_03196 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 53549; End: 55138 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 58 | 514 | 1.2e-120 | 0.9977777777777778 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 4.45e-22 | 65 | 513 | 42 | 425 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02057 | Glyco_hydro_59 | 1.40e-04 | 209 | 377 | 111 | 252 | Glycosyl hydrolase family 59. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT40981.1 | 1.60e-253 | 1 | 517 | 1 | 511 |
| QIU93337.1 | 1.18e-245 | 1 | 525 | 13 | 531 |
| BCA52347.1 | 2.60e-162 | 1 | 524 | 1 | 520 |
| QMW85450.1 | 2.73e-159 | 1 | 524 | 1 | 520 |
| AAO78132.1 | 2.73e-159 | 1 | 524 | 1 | 520 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6IUJ_A | 5.50e-13 | 58 | 424 | 23 | 384 | ChainA, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
| 6KRL_A | 6.51e-13 | 58 | 424 | 94 | 455 | Crystalstructure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris [Saccharomyces uvarum],6KRN_A Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid [Saccharomyces uvarum] |
| 3GTN_A | 6.84e-10 | 178 | 461 | 81 | 335 | CrystalStructure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3GTN_B Crystal Structure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3KL0_A Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_B Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_C Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_D Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL3_A Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_B Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_C Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_D Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL5_A Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_B Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_C Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_D Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q45070 | 4.02e-09 | 178 | 461 | 112 | 366 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| G5ECR8 | 1.75e-06 | 57 | 493 | 91 | 522 | Putative glucosylceramidase 3 OS=Caenorhabditis elegans OX=6239 GN=gba-3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
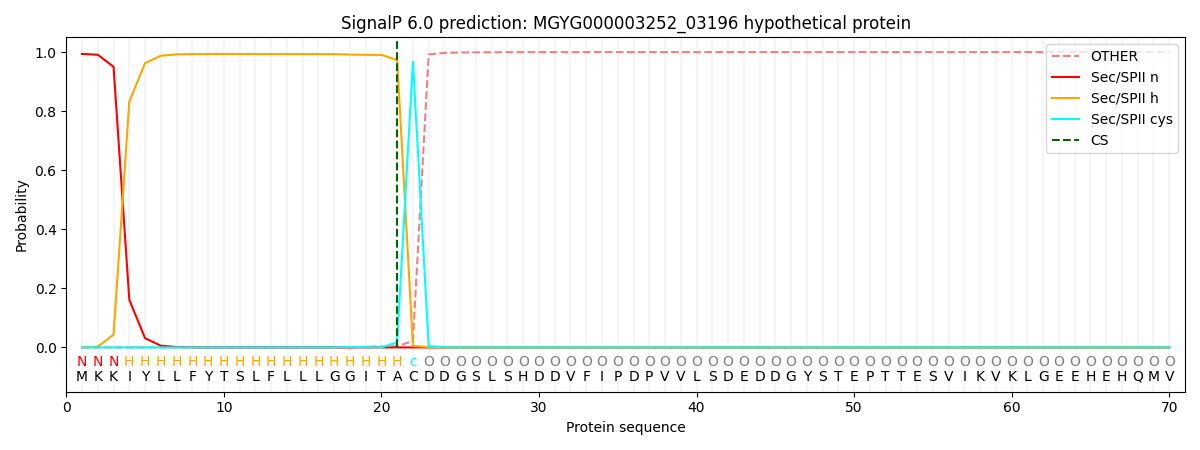
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000039 | 0.006005 | 0.993975 | 0.000004 | 0.000005 | 0.000004 |
